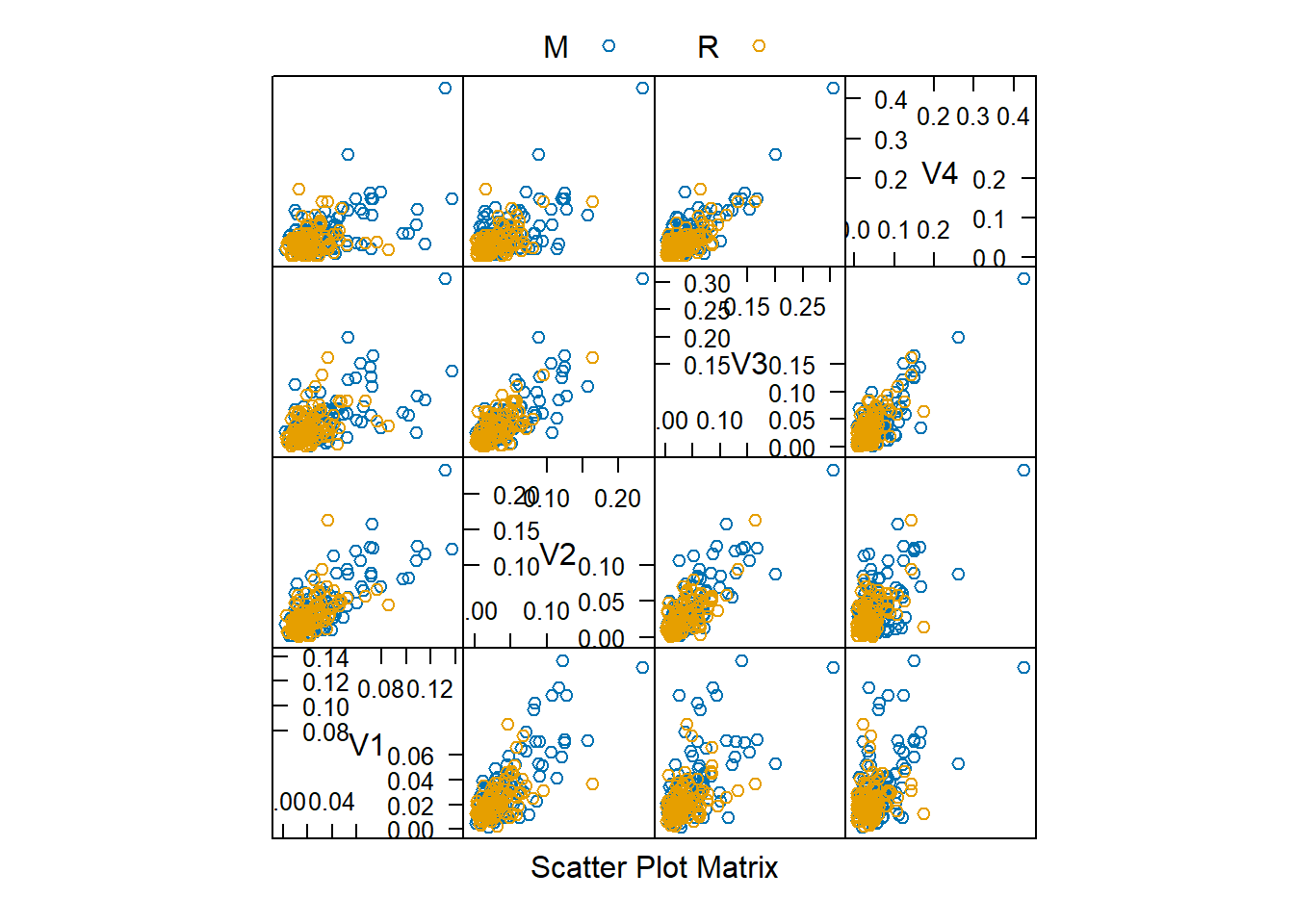
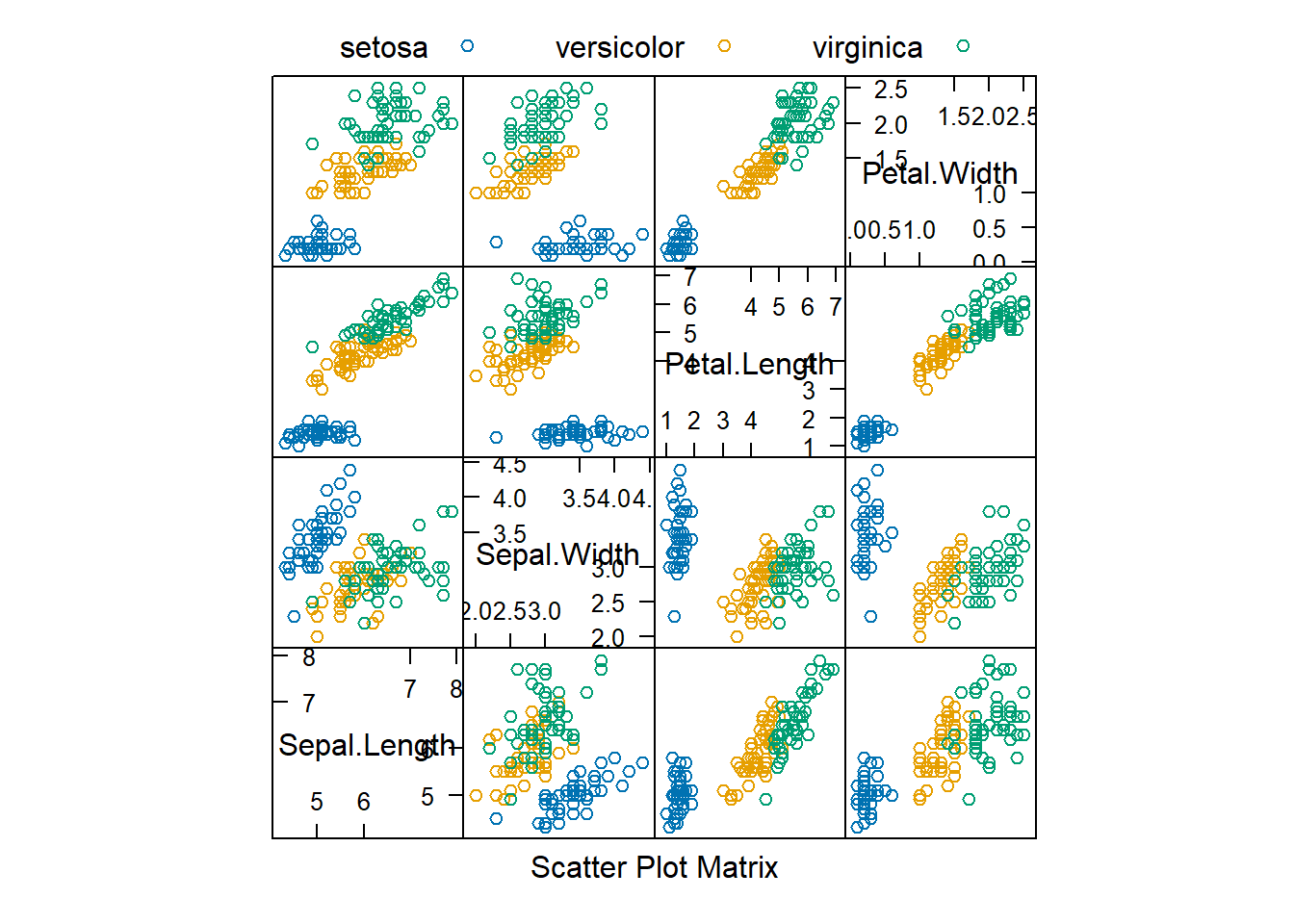
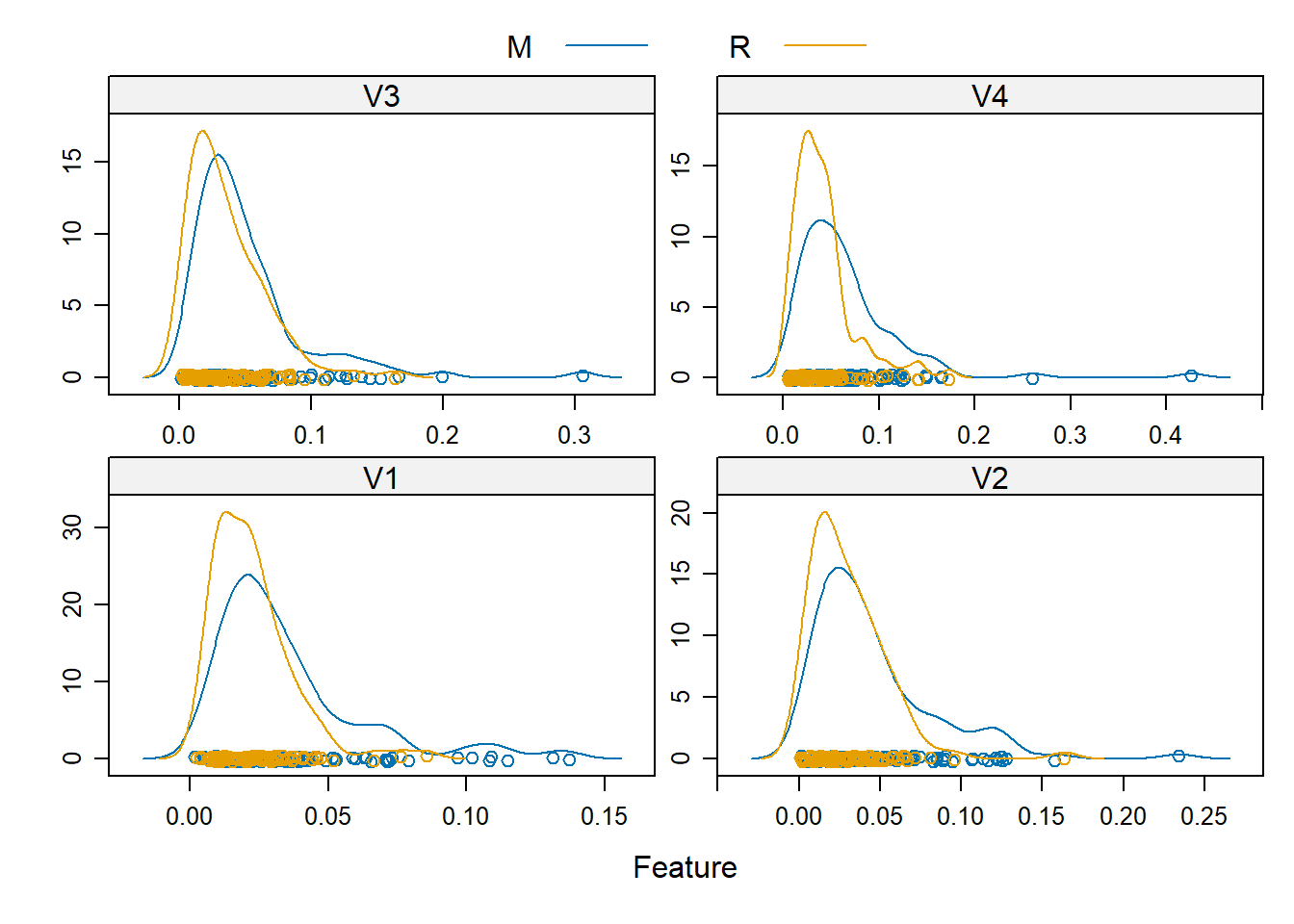
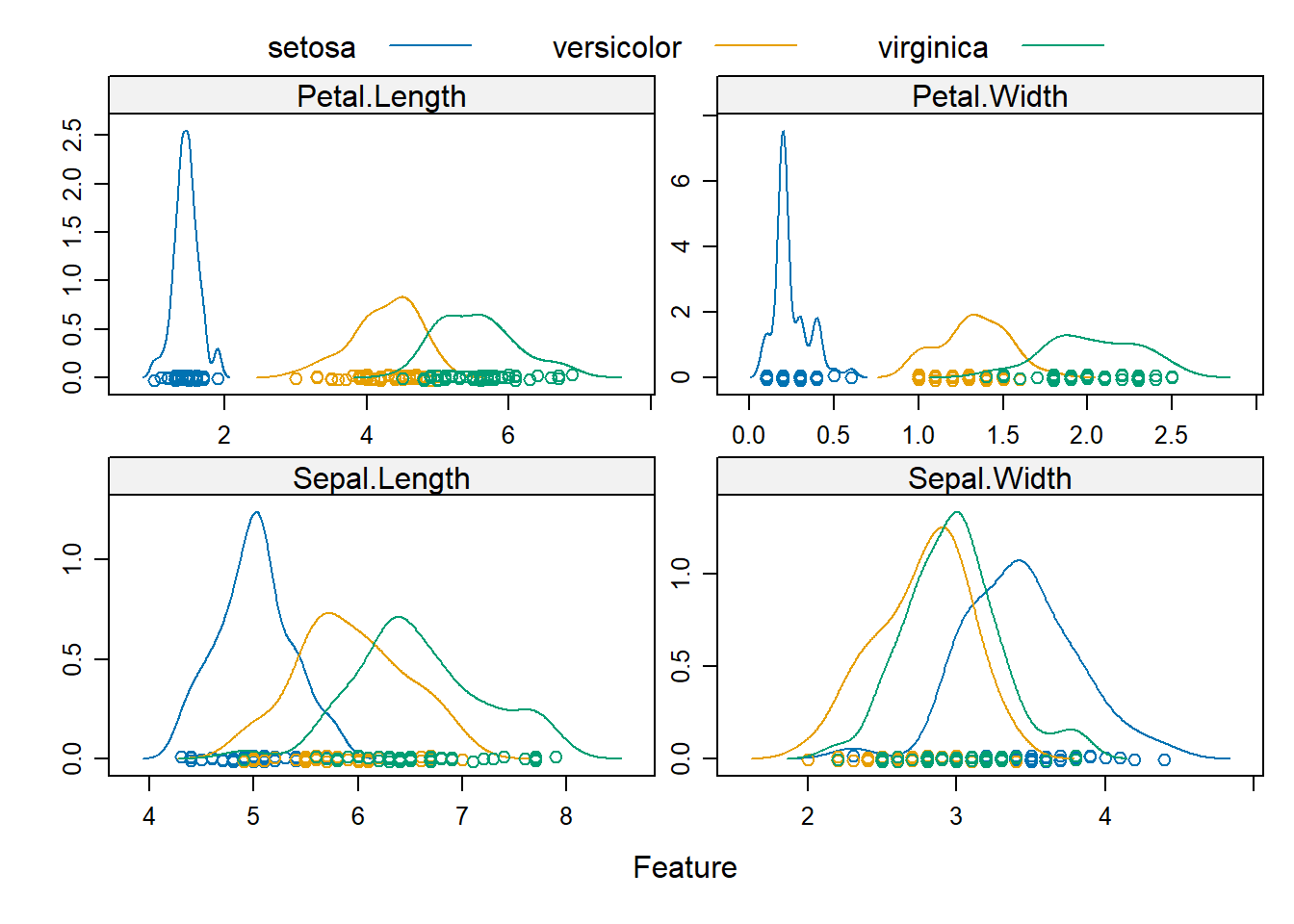
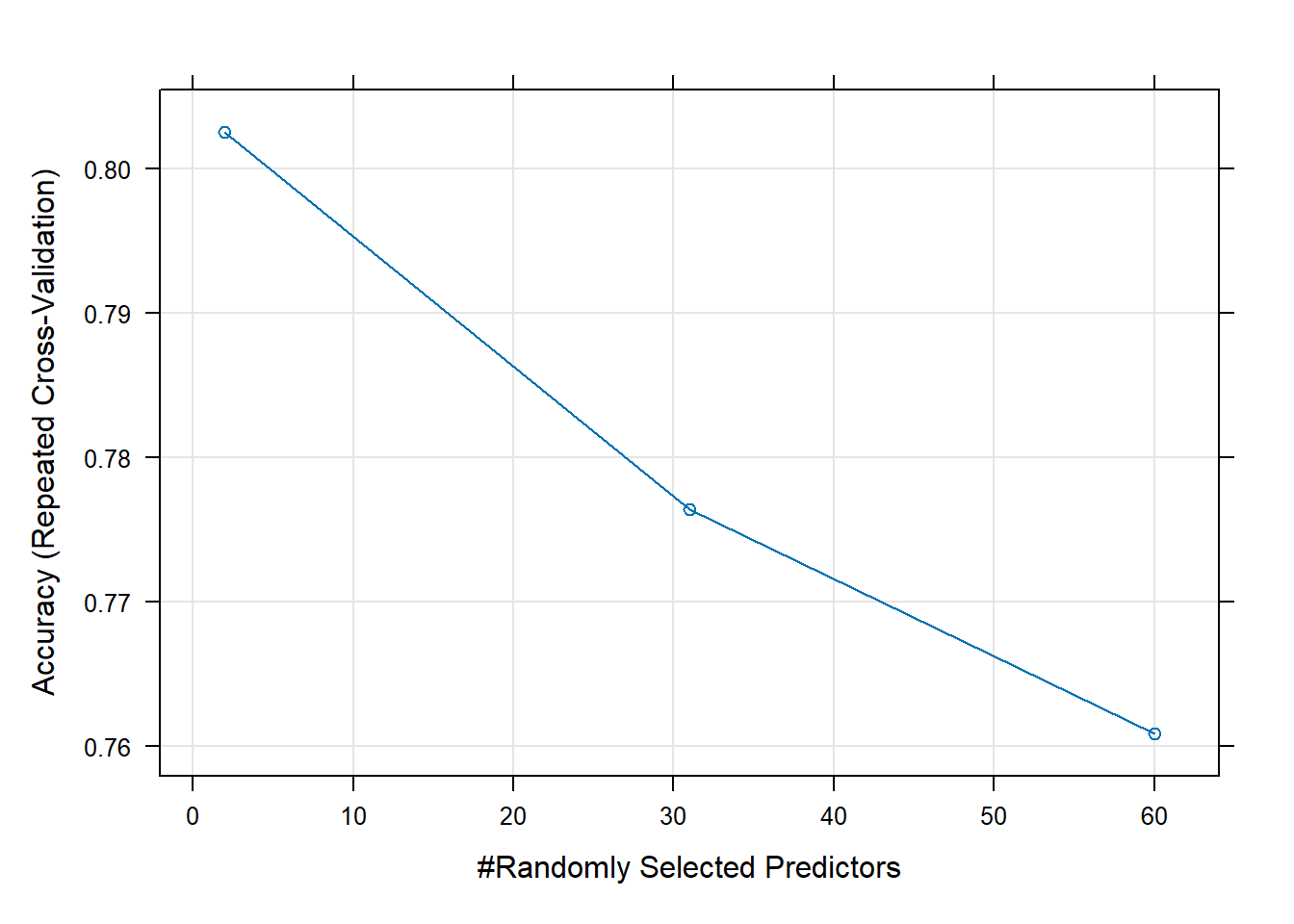
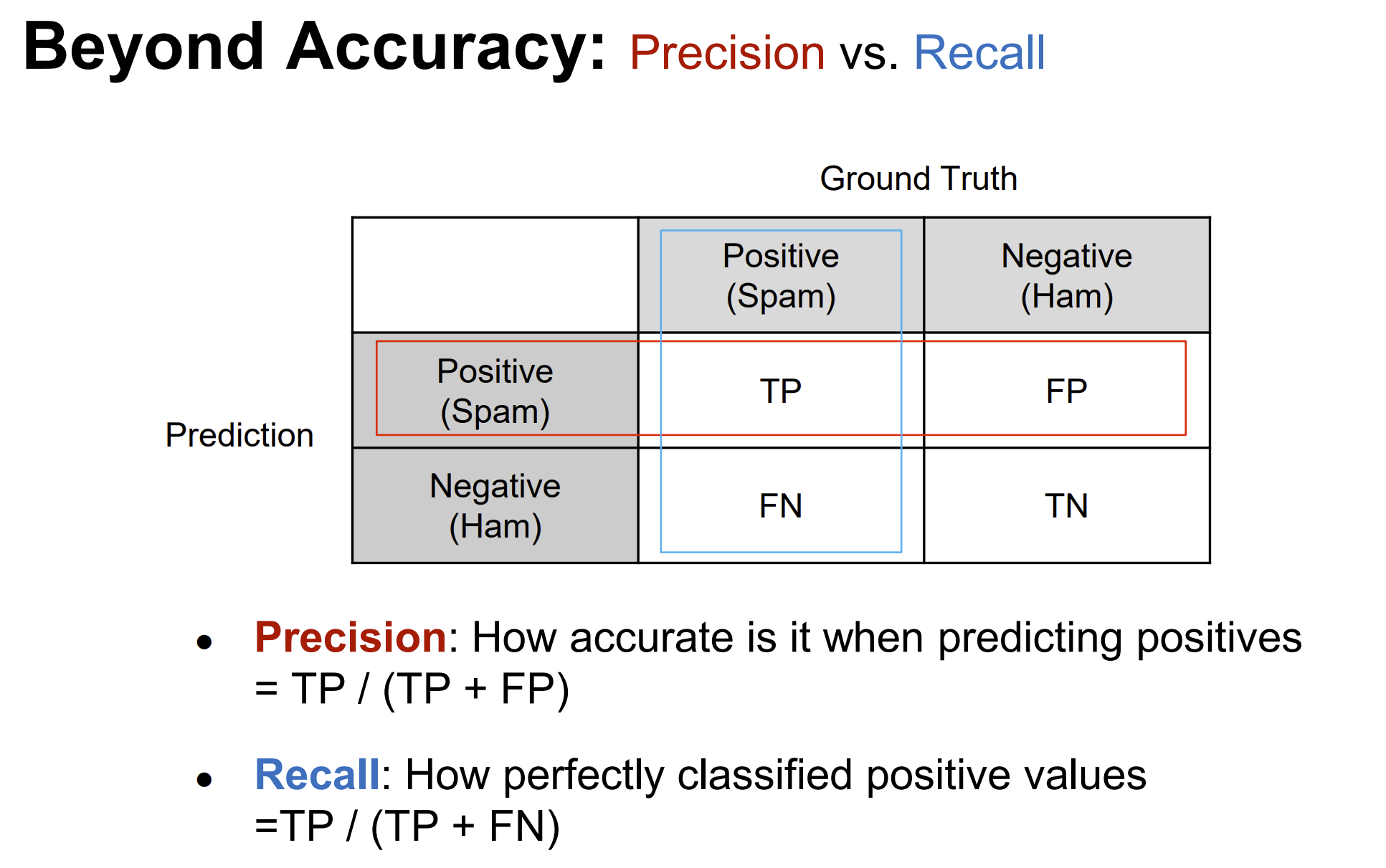
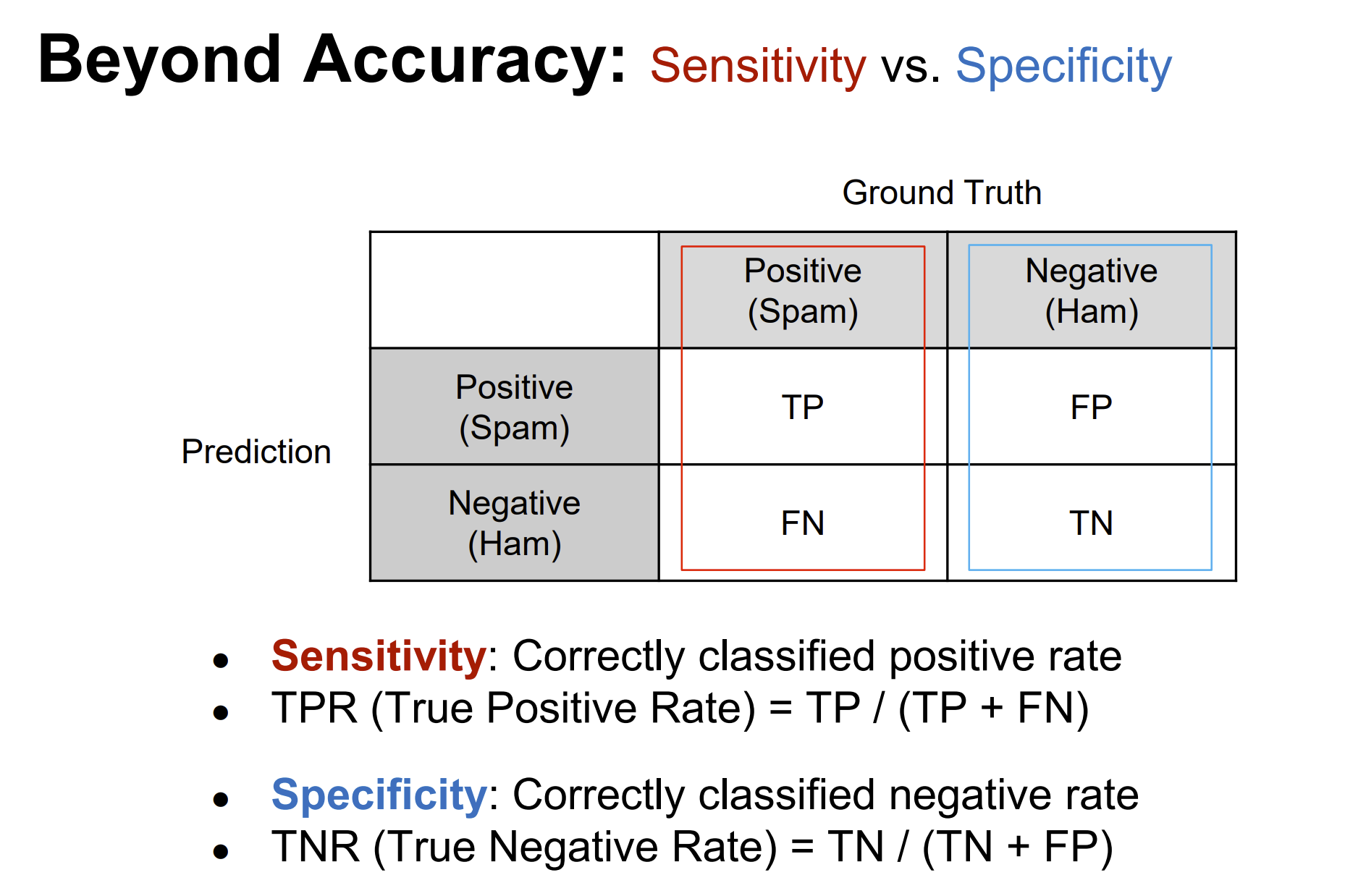
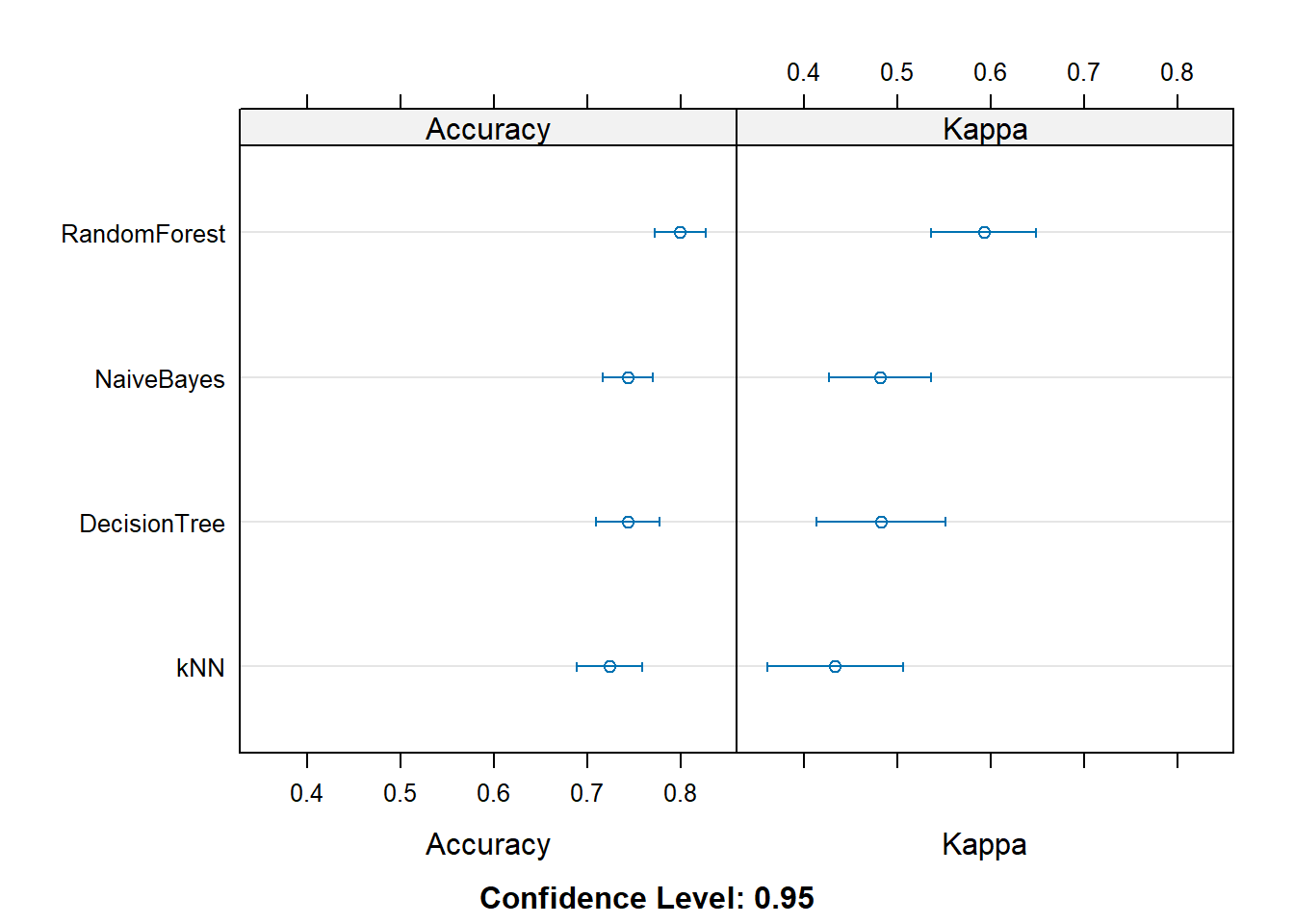
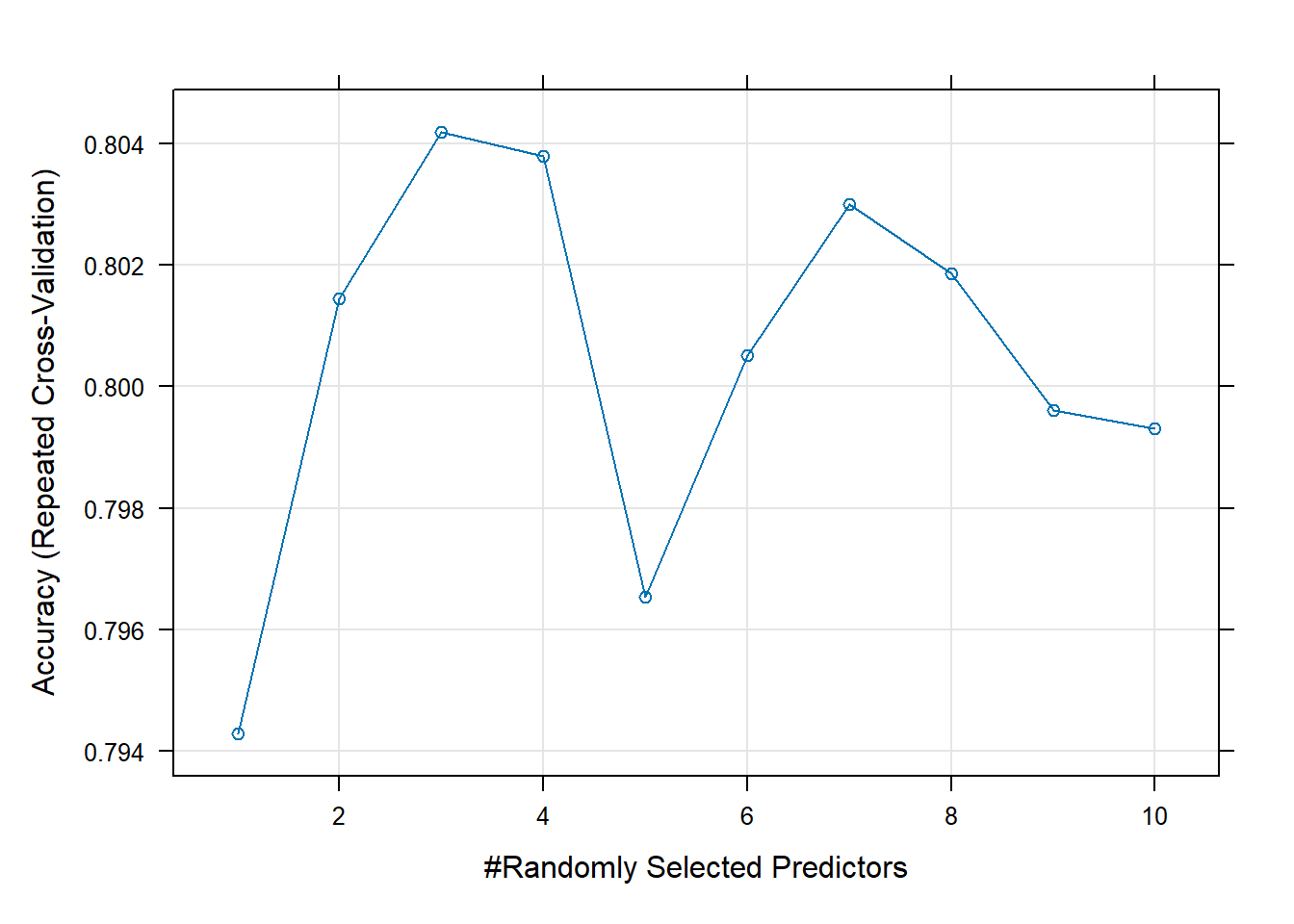
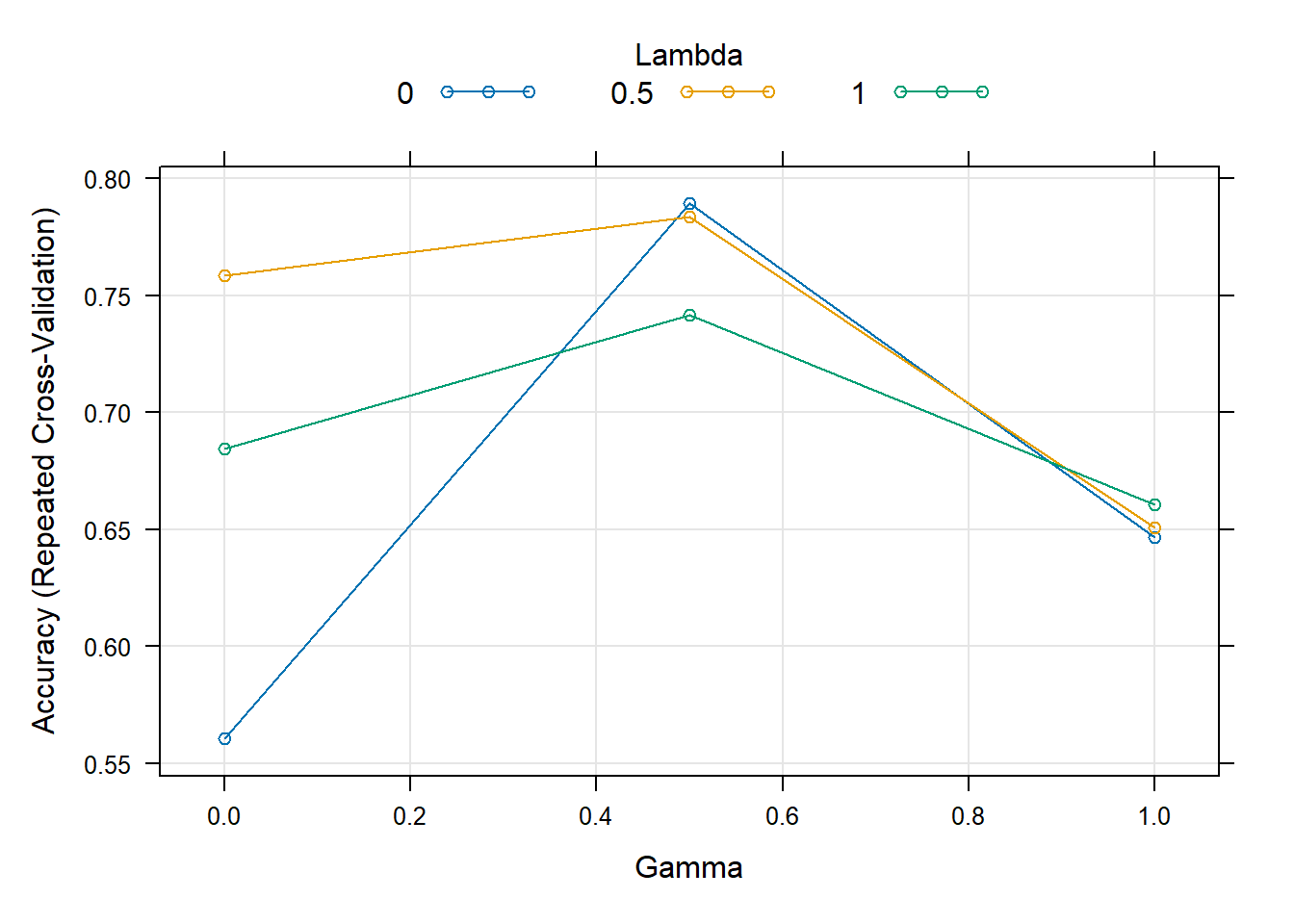
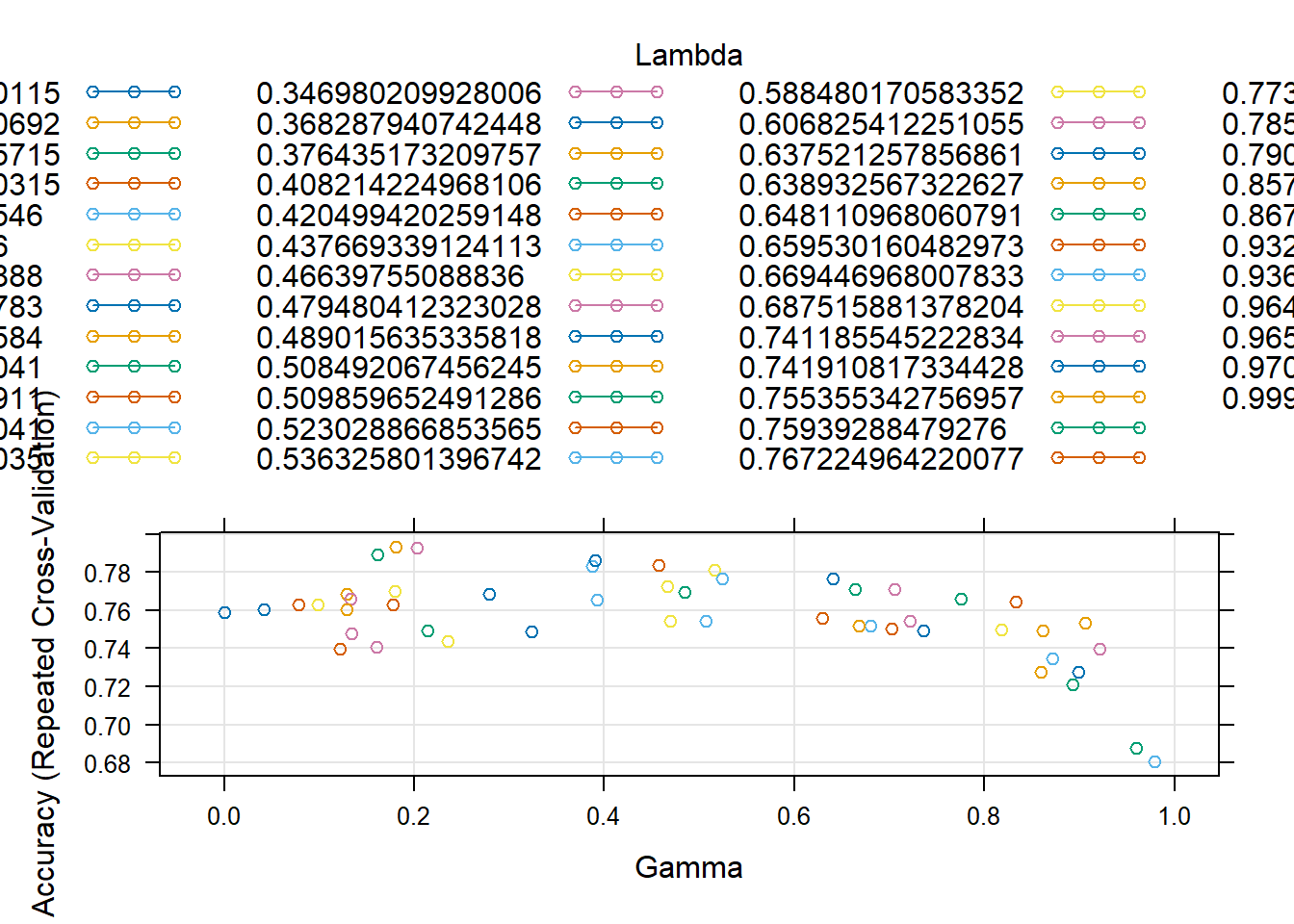
Rows: 208
Columns: 61
$ V1 <dbl> 0.0200, 0.0453, 0.0262, 0.0100, 0.0762, 0.0286, 0.0317, 0.0519, …
$ V2 <dbl> 0.0371, 0.0523, 0.0582, 0.0171, 0.0666, 0.0453, 0.0956, 0.0548, …
$ V3 <dbl> 0.0428, 0.0843, 0.1099, 0.0623, 0.0481, 0.0277, 0.1321, 0.0842, …
$ V4 <dbl> 0.0207, 0.0689, 0.1083, 0.0205, 0.0394, 0.0174, 0.1408, 0.0319, …
$ V5 <dbl> 0.0954, 0.1183, 0.0974, 0.0205, 0.0590, 0.0384, 0.1674, 0.1158, …
$ V6 <dbl> 0.0986, 0.2583, 0.2280, 0.0368, 0.0649, 0.0990, 0.1710, 0.0922, …
$ V7 <dbl> 0.1539, 0.2156, 0.2431, 0.1098, 0.1209, 0.1201, 0.0731, 0.1027, …
$ V8 <dbl> 0.1601, 0.3481, 0.3771, 0.1276, 0.2467, 0.1833, 0.1401, 0.0613, …
$ V9 <dbl> 0.3109, 0.3337, 0.5598, 0.0598, 0.3564, 0.2105, 0.2083, 0.1465, …
$ V10 <dbl> 0.2111, 0.2872, 0.6194, 0.1264, 0.4459, 0.3039, 0.3513, 0.2838, …
$ V11 <dbl> 0.1609, 0.4918, 0.6333, 0.0881, 0.4152, 0.2988, 0.1786, 0.2802, …
$ V12 <dbl> 0.1582, 0.6552, 0.7060, 0.1992, 0.3952, 0.4250, 0.0658, 0.3086, …
$ V13 <dbl> 0.2238, 0.6919, 0.5544, 0.0184, 0.4256, 0.6343, 0.0513, 0.2657, …
$ V14 <dbl> 0.0645, 0.7797, 0.5320, 0.2261, 0.4135, 0.8198, 0.3752, 0.3801, …
$ V15 <dbl> 0.0660, 0.7464, 0.6479, 0.1729, 0.4528, 1.0000, 0.5419, 0.5626, …
$ V16 <dbl> 0.2273, 0.9444, 0.6931, 0.2131, 0.5326, 0.9988, 0.5440, 0.4376, …
$ V17 <dbl> 0.3100, 1.0000, 0.6759, 0.0693, 0.7306, 0.9508, 0.5150, 0.2617, …
$ V18 <dbl> 0.2999, 0.8874, 0.7551, 0.2281, 0.6193, 0.9025, 0.4262, 0.1199, …
$ V19 <dbl> 0.5078, 0.8024, 0.8929, 0.4060, 0.2032, 0.7234, 0.2024, 0.6676, …
$ V20 <dbl> 0.4797, 0.7818, 0.8619, 0.3973, 0.4636, 0.5122, 0.4233, 0.9402, …
$ V21 <dbl> 0.5783, 0.5212, 0.7974, 0.2741, 0.4148, 0.2074, 0.7723, 0.7832, …
$ V22 <dbl> 0.5071, 0.4052, 0.6737, 0.3690, 0.4292, 0.3985, 0.9735, 0.5352, …
$ V23 <dbl> 0.4328, 0.3957, 0.4293, 0.5556, 0.5730, 0.5890, 0.9390, 0.6809, …
$ V24 <dbl> 0.5550, 0.3914, 0.3648, 0.4846, 0.5399, 0.2872, 0.5559, 0.9174, …
$ V25 <dbl> 0.6711, 0.3250, 0.5331, 0.3140, 0.3161, 0.2043, 0.5268, 0.7613, …
$ V26 <dbl> 0.6415, 0.3200, 0.2413, 0.5334, 0.2285, 0.5782, 0.6826, 0.8220, …
$ V27 <dbl> 0.7104, 0.3271, 0.5070, 0.5256, 0.6995, 0.5389, 0.5713, 0.8872, …
$ V28 <dbl> 0.8080, 0.2767, 0.8533, 0.2520, 1.0000, 0.3750, 0.5429, 0.6091, …
$ V29 <dbl> 0.6791, 0.4423, 0.6036, 0.2090, 0.7262, 0.3411, 0.2177, 0.2967, …
$ V30 <dbl> 0.3857, 0.2028, 0.8514, 0.3559, 0.4724, 0.5067, 0.2149, 0.1103, …
$ V31 <dbl> 0.1307, 0.3788, 0.8512, 0.6260, 0.5103, 0.5580, 0.5811, 0.1318, …
$ V32 <dbl> 0.2604, 0.2947, 0.5045, 0.7340, 0.5459, 0.4778, 0.6323, 0.0624, …
$ V33 <dbl> 0.5121, 0.1984, 0.1862, 0.6120, 0.2881, 0.3299, 0.2965, 0.0990, …
$ V34 <dbl> 0.7547, 0.2341, 0.2709, 0.3497, 0.0981, 0.2198, 0.1873, 0.4006, …
$ V35 <dbl> 0.8537, 0.1306, 0.4232, 0.3953, 0.1951, 0.1407, 0.2969, 0.3666, …
$ V36 <dbl> 0.8507, 0.4182, 0.3043, 0.3012, 0.4181, 0.2856, 0.5163, 0.1050, …
$ V37 <dbl> 0.6692, 0.3835, 0.6116, 0.5408, 0.4604, 0.3807, 0.6153, 0.1915, …
$ V38 <dbl> 0.6097, 0.1057, 0.6756, 0.8814, 0.3217, 0.4158, 0.4283, 0.3930, …
$ V39 <dbl> 0.4943, 0.1840, 0.5375, 0.9857, 0.2828, 0.4054, 0.5479, 0.4288, …
$ V40 <dbl> 0.2744, 0.1970, 0.4719, 0.9167, 0.2430, 0.3296, 0.6133, 0.2546, …
$ V41 <dbl> 0.0510, 0.1674, 0.4647, 0.6121, 0.1979, 0.2707, 0.5017, 0.1151, …
$ V42 <dbl> 0.2834, 0.0583, 0.2587, 0.5006, 0.2444, 0.2650, 0.2377, 0.2196, …
$ V43 <dbl> 0.2825, 0.1401, 0.2129, 0.3210, 0.1847, 0.0723, 0.1957, 0.1879, …
$ V44 <dbl> 0.4256, 0.1628, 0.2222, 0.3202, 0.0841, 0.1238, 0.1749, 0.1437, …
$ V45 <dbl> 0.2641, 0.0621, 0.2111, 0.4295, 0.0692, 0.1192, 0.1304, 0.2146, …
$ V46 <dbl> 0.1386, 0.0203, 0.0176, 0.3654, 0.0528, 0.1089, 0.0597, 0.2360, …
$ V47 <dbl> 0.1051, 0.0530, 0.1348, 0.2655, 0.0357, 0.0623, 0.1124, 0.1125, …
$ V48 <dbl> 0.1343, 0.0742, 0.0744, 0.1576, 0.0085, 0.0494, 0.1047, 0.0254, …
$ V49 <dbl> 0.0383, 0.0409, 0.0130, 0.0681, 0.0230, 0.0264, 0.0507, 0.0285, …
$ V50 <dbl> 0.0324, 0.0061, 0.0106, 0.0294, 0.0046, 0.0081, 0.0159, 0.0178, …
$ V51 <dbl> 0.0232, 0.0125, 0.0033, 0.0241, 0.0156, 0.0104, 0.0195, 0.0052, …
$ V52 <dbl> 0.0027, 0.0084, 0.0232, 0.0121, 0.0031, 0.0045, 0.0201, 0.0081, …
$ V53 <dbl> 0.0065, 0.0089, 0.0166, 0.0036, 0.0054, 0.0014, 0.0248, 0.0120, …
$ V54 <dbl> 0.0159, 0.0048, 0.0095, 0.0150, 0.0105, 0.0038, 0.0131, 0.0045, …
$ V55 <dbl> 0.0072, 0.0094, 0.0180, 0.0085, 0.0110, 0.0013, 0.0070, 0.0121, …
$ V56 <dbl> 0.0167, 0.0191, 0.0244, 0.0073, 0.0015, 0.0089, 0.0138, 0.0097, …
$ V57 <dbl> 0.0180, 0.0140, 0.0316, 0.0050, 0.0072, 0.0057, 0.0092, 0.0085, …
$ V58 <dbl> 0.0084, 0.0049, 0.0164, 0.0044, 0.0048, 0.0027, 0.0143, 0.0047, …
$ V59 <dbl> 0.0090, 0.0052, 0.0095, 0.0040, 0.0107, 0.0051, 0.0036, 0.0048, …
$ V60 <dbl> 0.0032, 0.0044, 0.0078, 0.0117, 0.0094, 0.0062, 0.0103, 0.0053, …
$ Class <fct> R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R, R…