Weekly design
Pre-class video
Data wrangling
VIDEO
# 02 Data processing using Base R # library (gapminder)library (dplyr)
Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Rows: 1,704
Columns: 6
$ country <fct> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", …
$ continent <fct> Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, …
$ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, …
$ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.8…
$ pop <int> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12…
$ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, …
c ("country" , "lifeExp" )]
# A tibble: 1,704 × 2
country lifeExp
<fct> <dbl>
1 Afghanistan 28.8
2 Afghanistan 30.3
3 Afghanistan 32.0
4 Afghanistan 34.0
5 Afghanistan 36.1
6 Afghanistan 38.4
7 Afghanistan 39.9
8 Afghanistan 40.8
9 Afghanistan 41.7
10 Afghanistan 41.8
# ℹ 1,694 more rows
c ("country" , "lifeExp" , "year" )]
# A tibble: 1,704 × 3
country lifeExp year
<fct> <dbl> <int>
1 Afghanistan 28.8 1952
2 Afghanistan 30.3 1957
3 Afghanistan 32.0 1962
4 Afghanistan 34.0 1967
5 Afghanistan 36.1 1972
6 Afghanistan 38.4 1977
7 Afghanistan 39.9 1982
8 Afghanistan 40.8 1987
9 Afghanistan 41.7 1992
10 Afghanistan 41.8 1997
# ℹ 1,694 more rows
# A tibble: 15 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
11 Afghanistan Asia 2002 42.1 25268405 727.
12 Afghanistan Asia 2007 43.8 31889923 975.
13 Albania Europe 1952 55.2 1282697 1601.
14 Albania Europe 1957 59.3 1476505 1942.
15 Albania Europe 1962 64.8 1728137 2313.
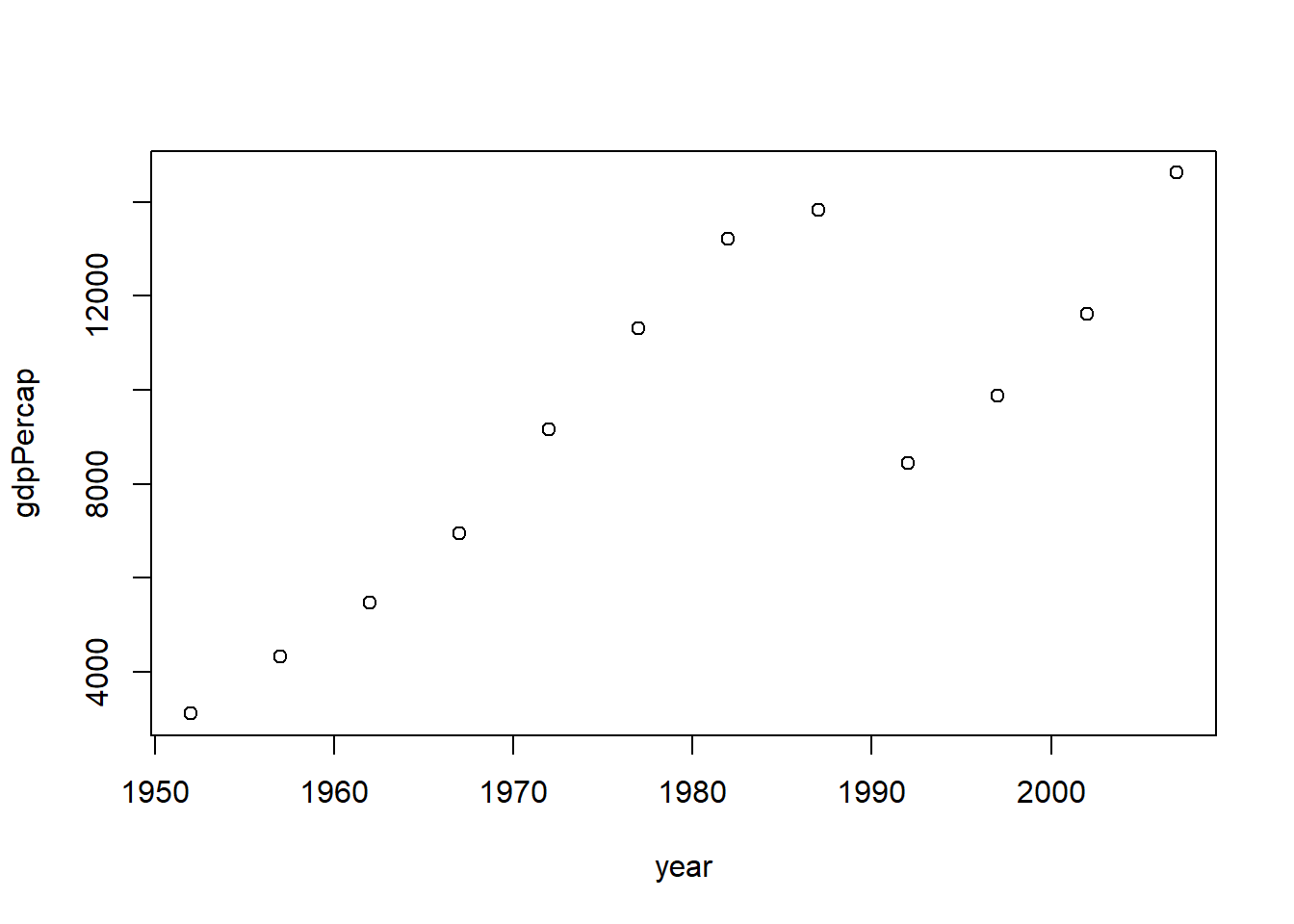
library (dplyr)%>% filter (country== "Croatia" ) %>% select (year, gdpPercap) %>% plot$ country == "Croatia" , ]
# A tibble: 12 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Croatia Europe 1952 61.2 3882229 3119.
2 Croatia Europe 1957 64.8 3991242 4338.
3 Croatia Europe 1962 67.1 4076557 5478.
4 Croatia Europe 1967 68.5 4174366 6960.
5 Croatia Europe 1972 69.6 4225310 9164.
6 Croatia Europe 1977 70.6 4318673 11305.
7 Croatia Europe 1982 70.5 4413368 13222.
8 Croatia Europe 1987 71.5 4484310 13823.
9 Croatia Europe 1992 72.5 4494013 8448.
10 Croatia Europe 1997 73.7 4444595 9876.
11 Croatia Europe 2002 74.9 4481020 11628.
12 Croatia Europe 2007 75.7 4493312 14619.
$ country == "Korea, Rep." , ]
# A tibble: 12 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Korea, Rep. Asia 1952 47.5 20947571 1031.
2 Korea, Rep. Asia 1957 52.7 22611552 1488.
3 Korea, Rep. Asia 1962 55.3 26420307 1536.
4 Korea, Rep. Asia 1967 57.7 30131000 2029.
5 Korea, Rep. Asia 1972 62.6 33505000 3031.
6 Korea, Rep. Asia 1977 64.8 36436000 4657.
7 Korea, Rep. Asia 1982 67.1 39326000 5623.
8 Korea, Rep. Asia 1987 69.8 41622000 8533.
9 Korea, Rep. Asia 1992 72.2 43805450 12104.
10 Korea, Rep. Asia 1997 74.6 46173816 15994.
11 Korea, Rep. Asia 2002 77.0 47969150 19234.
12 Korea, Rep. Asia 2007 78.6 49044790 23348.
levels (gapminder$ country)
[1] "Afghanistan" "Albania"
[3] "Algeria" "Angola"
[5] "Argentina" "Australia"
[7] "Austria" "Bahrain"
[9] "Bangladesh" "Belgium"
[11] "Benin" "Bolivia"
[13] "Bosnia and Herzegovina" "Botswana"
[15] "Brazil" "Bulgaria"
[17] "Burkina Faso" "Burundi"
[19] "Cambodia" "Cameroon"
[21] "Canada" "Central African Republic"
[23] "Chad" "Chile"
[25] "China" "Colombia"
[27] "Comoros" "Congo, Dem. Rep."
[29] "Congo, Rep." "Costa Rica"
[31] "Cote d'Ivoire" "Croatia"
[33] "Cuba" "Czech Republic"
[35] "Denmark" "Djibouti"
[37] "Dominican Republic" "Ecuador"
[39] "Egypt" "El Salvador"
[41] "Equatorial Guinea" "Eritrea"
[43] "Ethiopia" "Finland"
[45] "France" "Gabon"
[47] "Gambia" "Germany"
[49] "Ghana" "Greece"
[51] "Guatemala" "Guinea"
[53] "Guinea-Bissau" "Haiti"
[55] "Honduras" "Hong Kong, China"
[57] "Hungary" "Iceland"
[59] "India" "Indonesia"
[61] "Iran" "Iraq"
[63] "Ireland" "Israel"
[65] "Italy" "Jamaica"
[67] "Japan" "Jordan"
[69] "Kenya" "Korea, Dem. Rep."
[71] "Korea, Rep." "Kuwait"
[73] "Lebanon" "Lesotho"
[75] "Liberia" "Libya"
[77] "Madagascar" "Malawi"
[79] "Malaysia" "Mali"
[81] "Mauritania" "Mauritius"
[83] "Mexico" "Mongolia"
[85] "Montenegro" "Morocco"
[87] "Mozambique" "Myanmar"
[89] "Namibia" "Nepal"
[91] "Netherlands" "New Zealand"
[93] "Nicaragua" "Niger"
[95] "Nigeria" "Norway"
[97] "Oman" "Pakistan"
[99] "Panama" "Paraguay"
[101] "Peru" "Philippines"
[103] "Poland" "Portugal"
[105] "Puerto Rico" "Reunion"
[107] "Romania" "Rwanda"
[109] "Sao Tome and Principe" "Saudi Arabia"
[111] "Senegal" "Serbia"
[113] "Sierra Leone" "Singapore"
[115] "Slovak Republic" "Slovenia"
[117] "Somalia" "South Africa"
[119] "Spain" "Sri Lanka"
[121] "Sudan" "Swaziland"
[123] "Sweden" "Switzerland"
[125] "Syria" "Taiwan"
[127] "Tanzania" "Thailand"
[129] "Togo" "Trinidad and Tobago"
[131] "Tunisia" "Turkey"
[133] "Uganda" "United Kingdom"
[135] "United States" "Uruguay"
[137] "Venezuela" "Vietnam"
[139] "West Bank and Gaza" "Yemen, Rep."
[141] "Zambia" "Zimbabwe"
$ country == "Croatia" , "pop" ]
# A tibble: 12 × 1
pop
<int>
1 3882229
2 3991242
3 4076557
4 4174366
5 4225310
6 4318673
7 4413368
8 4484310
9 4494013
10 4444595
11 4481020
12 4493312
$ country == "Croatia" , c ("lifeExp" ,"pop" )]
# A tibble: 12 × 2
lifeExp pop
<dbl> <int>
1 61.2 3882229
2 64.8 3991242
3 67.1 4076557
4 68.5 4174366
5 69.6 4225310
6 70.6 4318673
7 70.5 4413368
8 71.5 4484310
9 72.5 4494013
10 73.7 4444595
11 74.9 4481020
12 75.7 4493312
$ country == "Croatia" & #Croatia extraction $ year > 1990 , #1990 after c ("lifeExp" ,"pop" )] # those variables
# A tibble: 4 × 2
lifeExp pop
<dbl> <int>
1 72.5 4494013
2 73.7 4444595
3 74.9 4481020
4 75.7 4493312
apply (gapminder[gapminder$ country == "Croatia" ,c ("lifeExp" ,"pop" )],2 , mean)
lifeExp pop
7.005592e+01 4.289916e+06
apply (gapminder[gapminder$ country == "Korea, Rep." ,c ("lifeExp" ,"pop" )],2 , mean)
lifeExp pop
65.001 36499386.333
# 03 Data processing using the dplyr library # select (gapminder, country, year, lifeExp)
# A tibble: 1,704 × 3
country year lifeExp
<fct> <int> <dbl>
1 Afghanistan 1952 28.8
2 Afghanistan 1957 30.3
3 Afghanistan 1962 32.0
4 Afghanistan 1967 34.0
5 Afghanistan 1972 36.1
6 Afghanistan 1977 38.4
7 Afghanistan 1982 39.9
8 Afghanistan 1987 40.8
9 Afghanistan 1992 41.7
10 Afghanistan 1997 41.8
# ℹ 1,694 more rows
filter (gapminder, country == "Croatia" )
# A tibble: 12 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Croatia Europe 1952 61.2 3882229 3119.
2 Croatia Europe 1957 64.8 3991242 4338.
3 Croatia Europe 1962 67.1 4076557 5478.
4 Croatia Europe 1967 68.5 4174366 6960.
5 Croatia Europe 1972 69.6 4225310 9164.
6 Croatia Europe 1977 70.6 4318673 11305.
7 Croatia Europe 1982 70.5 4413368 13222.
8 Croatia Europe 1987 71.5 4484310 13823.
9 Croatia Europe 1992 72.5 4494013 8448.
10 Croatia Europe 1997 73.7 4444595 9876.
11 Croatia Europe 2002 74.9 4481020 11628.
12 Croatia Europe 2007 75.7 4493312 14619.
summarize (gapminder, pop_avg = mean (pop))
# A tibble: 1 × 1
pop_avg
<dbl>
1 29601212.
summarize (group_by (gapminder, continent), pop_avg = mean (pop))
# A tibble: 5 × 2
continent pop_avg
<fct> <dbl>
1 Africa 9916003.
2 Americas 24504795.
3 Asia 77038722.
4 Europe 17169765.
5 Oceania 8874672.
summarize (group_by (gapminder, continent, country), pop_avg = mean (pop))
`summarise()` has grouped output by 'continent'. You can override using the
`.groups` argument.
# A tibble: 142 × 3
# Groups: continent [5]
continent country pop_avg
<fct> <fct> <dbl>
1 Africa Algeria 19875406.
2 Africa Angola 7309390.
3 Africa Benin 4017497.
4 Africa Botswana 971186.
5 Africa Burkina Faso 7548677.
6 Africa Burundi 4651608.
7 Africa Cameroon 9816648.
8 Africa Central African Republic 2560963
9 Africa Chad 5329256.
10 Africa Comoros 361684.
# ℹ 132 more rows
%>% group_by (continent, country) %>% summarize (pop_avg = mean (pop))
`summarise()` has grouped output by 'continent'. You can override using the
`.groups` argument.
# A tibble: 142 × 3
# Groups: continent [5]
continent country pop_avg
<fct> <fct> <dbl>
1 Africa Algeria 19875406.
2 Africa Angola 7309390.
3 Africa Benin 4017497.
4 Africa Botswana 971186.
5 Africa Burkina Faso 7548677.
6 Africa Burundi 4651608.
7 Africa Cameroon 9816648.
8 Africa Central African Republic 2560963
9 Africa Chad 5329256.
10 Africa Comoros 361684.
# ℹ 132 more rows
= filter (gapminder, country == "Croatia" )= select (temp1, country, year, lifeExp)= apply (temp2[ , c ("lifeExp" )], 2 , mean)
%>% filter (country == "Croatia" ) %>% select (country, year, lifeExp) %>% summarize (lifeExp_avg = mean (lifeExp))
# A tibble: 1 × 1
lifeExp_avg
<dbl>
1 70.1
data in need: avocado.csv
# 04 The reality of data processing # library (ggplot2)<- read.csv ("data/avocado.csv" , header= TRUE , sep = "," )str (avocado)
'data.frame': 18249 obs. of 14 variables:
$ X : int 0 1 2 3 4 5 6 7 8 9 ...
$ Date : chr "2015-12-27" "2015-12-20" "2015-12-13" "2015-12-06" ...
$ AveragePrice: num 1.33 1.35 0.93 1.08 1.28 1.26 0.99 0.98 1.02 1.07 ...
$ Total.Volume: num 64237 54877 118220 78992 51040 ...
$ X4046 : num 1037 674 795 1132 941 ...
$ X4225 : num 54455 44639 109150 71976 43838 ...
$ X4770 : num 48.2 58.3 130.5 72.6 75.8 ...
$ Total.Bags : num 8697 9506 8145 5811 6184 ...
$ Small.Bags : num 8604 9408 8042 5677 5986 ...
$ Large.Bags : num 93.2 97.5 103.1 133.8 197.7 ...
$ XLarge.Bags : num 0 0 0 0 0 0 0 0 0 0 ...
$ type : chr "conventional" "conventional" "conventional" "conventional" ...
$ year : int 2015 2015 2015 2015 2015 2015 2015 2015 2015 2015 ...
$ region : chr "Albany" "Albany" "Albany" "Albany" ...
x_avg = avocado %>% group_by (region) %>% summarize (V_avg = mean (Total.Volume), P_avg = mean (AveragePrice)))
# A tibble: 54 × 3
region V_avg P_avg
<chr> <dbl> <dbl>
1 Albany 47538. 1.56
2 Atlanta 262145. 1.34
3 BaltimoreWashington 398562. 1.53
4 Boise 42643. 1.35
5 Boston 287793. 1.53
6 BuffaloRochester 67936. 1.52
7 California 3044324. 1.40
8 Charlotte 105194. 1.61
9 Chicago 395569. 1.56
10 CincinnatiDayton 131722. 1.21
# ℹ 44 more rows
x_avg = avocado %>% group_by (region, year) %>% summarize (V_avg = mean (Total.Volume), P_avg = mean (AveragePrice)))
`summarise()` has grouped output by 'region'. You can override using the
`.groups` argument.
# A tibble: 216 × 4
# Groups: region [54]
region year V_avg P_avg
<chr> <int> <dbl> <dbl>
1 Albany 2015 38749. 1.54
2 Albany 2016 50619. 1.53
3 Albany 2017 49355. 1.64
4 Albany 2018 64249. 1.44
5 Atlanta 2015 223382. 1.38
6 Atlanta 2016 272374. 1.21
7 Atlanta 2017 271841. 1.43
8 Atlanta 2018 342976. 1.29
9 BaltimoreWashington 2015 390823. 1.37
10 BaltimoreWashington 2016 393210. 1.59
# ℹ 206 more rows
= avocado %>% group_by (region, year, type) %>% summarize (V_avg = mean (Total.Volume), P_avg = mean (AveragePrice))
`summarise()` has grouped output by 'region', 'year'. You can override using
the `.groups` argument.
%>% group_by (region, year, type) %>% summarize (V_avg = mean (Total.Volume),P_avg = mean (AveragePrice)) -> x_avg
`summarise()` has grouped output by 'region', 'year'. You can override using
the `.groups` argument.
%>% filter (region != "TotalUS" ) %>% ggplot (aes (year, V_avg, col = type)) + geom_line () + facet_wrap (~ region)
# install.packages("ggplot2") library (ggplot2)arrange (x_avg, desc (V_avg))
# A tibble: 432 × 5
# Groups: region, year [216]
region year type V_avg P_avg
<chr> <int> <chr> <dbl> <dbl>
1 TotalUS 2018 conventional 42125533. 1.06
2 TotalUS 2016 conventional 34043450. 1.05
3 TotalUS 2017 conventional 33995658. 1.22
4 TotalUS 2015 conventional 31224729. 1.01
5 SouthCentral 2018 conventional 7465557. 0.806
6 West 2018 conventional 7451445. 0.981
7 California 2018 conventional 6786962. 1.08
8 West 2016 conventional 6404892. 0.916
9 West 2017 conventional 6279482. 1.10
10 California 2016 conventional 6105539. 1.05
# ℹ 422 more rows
= x_avg %>% filter (region != "TotalUS" )
# After excluding TotalUS, you can process it using statistical functions directly. $ V_avg == max (x_avg1$ V_avg),]
# A tibble: 1 × 5
# Groups: region, year [1]
region year type V_avg P_avg
<chr> <int> <chr> <dbl> <dbl>
1 SouthCentral 2018 conventional 7465557. 0.806
# install.packages("lubridate") library (lubridate)
Attaching package: 'lubridate'
The following objects are masked from 'package:base':
date, intersect, setdiff, union
x_avg = avocado %>% group_by (region, year, month (Date), type) %>% summarize (V_avg = mean (Total.Volume), P_avg = mean (AveragePrice)))
`summarise()` has grouped output by 'region', 'year', 'month(Date)'. You can
override using the `.groups` argument.
# A tibble: 4,212 × 6
# Groups: region, year, month(Date) [2,106]
region year `month(Date)` type V_avg P_avg
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Albany 2015 1 conventional 42932. 1.17
2 Albany 2015 1 organic 1198. 1.84
3 Albany 2015 2 conventional 52343. 1.03
4 Albany 2015 2 organic 1334. 1.76
5 Albany 2015 3 conventional 50659. 1.06
6 Albany 2015 3 organic 1444. 1.83
7 Albany 2015 4 conventional 48594. 1.17
8 Albany 2015 4 organic 1402. 1.89
9 Albany 2015 5 conventional 97216. 1.26
10 Albany 2015 5 organic 1836. 1.94
# ℹ 4,202 more rows
Class
Introduction to Tidyverse
The tidyverse is a powerful collection of R packages that are actually data tools for transforming and visualizing data. All packages of the tidyverse share an underlying philosophy and common APls.
The core packages are:
ggplot2, which implements the grammar of graphics. You can use it to visualize your data.
dplyr is a grammar of data You can use it to solve the most common data manipulation challenges.
tidyr helps you to create tidy data or data where each variable is in a column, each observation is a row end each value is a column, each observation is a row end each value is a cell.
readr is a fast and friendly way to read rectangular
purrr enhances R’s functional programming (FP)toolkit by providing a complete and consistent set of tools for working with functions and vectors.
tibble is a modern re-imaginging of the data
stringr provides a cohesive set of functions designed to make working with strings as easy as possible
forcats provide a suite of useful tools that solve common problems with factors.
tidyverse
Before Tidyverse Before the tidyverse, R programming was largely centered around base R functions and packages. This included using base R functions for data manipulation (like subsetmergeapplyplothist
After Tidyverse The tidyverse, developed by Hadley Wickham and others, brought a suite of packages designed to work harmoniously together using a consistent syntax and underlying philosophy. Key features and impacts include:
Consistent Syntax : The tidyverse introduced a consistent and readable syntax that leverages chaining operations using the %>%magrittr
Data Manipulation : With dplyrfilter()arrange()select()mutate()summarise()
Data Importing and Tidying : readrtidyr
Visualization : ggplot2
Community and Accessibility : The tidyverse has fostered a strong community and has contributed significantly to teaching materials that are user-friendly and accessible to beginners. This has democratized data analysis in R, making it more accessible to non-programmers.
Impact on Package Development : The tidyverse’s philosophy and popularity have influenced the development of other packages, even those not part of the tidyverse, to adopt tidy principles and interoperate smoothly with tidyverse packages.
The tidyverse has not only changed the syntax and functionality of R coding but also its philosophy towards data analysis. It promotes a workflow that is coherent, transparent, and efficient, which has been widely adopted in academia, industry, and teaching. While some veteran R users prefer the flexibility and control of base R, the tidyverse’s approachable syntax and powerful capabilities have made it a pivotal tool in modern R programming, particularly for data science.
Please check out the homepage of tidyverse: https://www.tidyverse.org/
You can install the complete tidyverse with:
> install.packages(“tidyverse”)
Then, load the core tidyverse and make it available in your current R session by running:
> library(tidyverse)
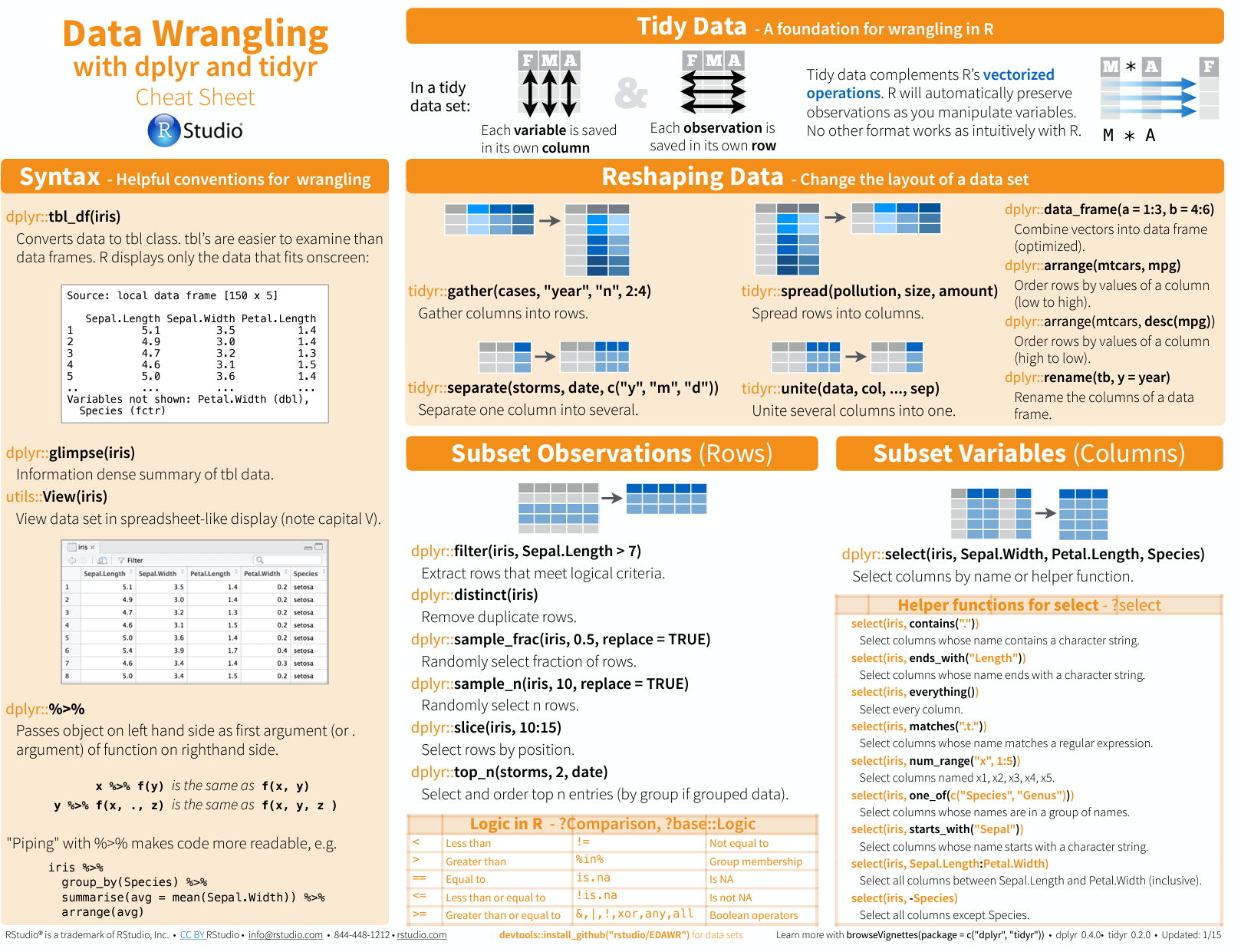
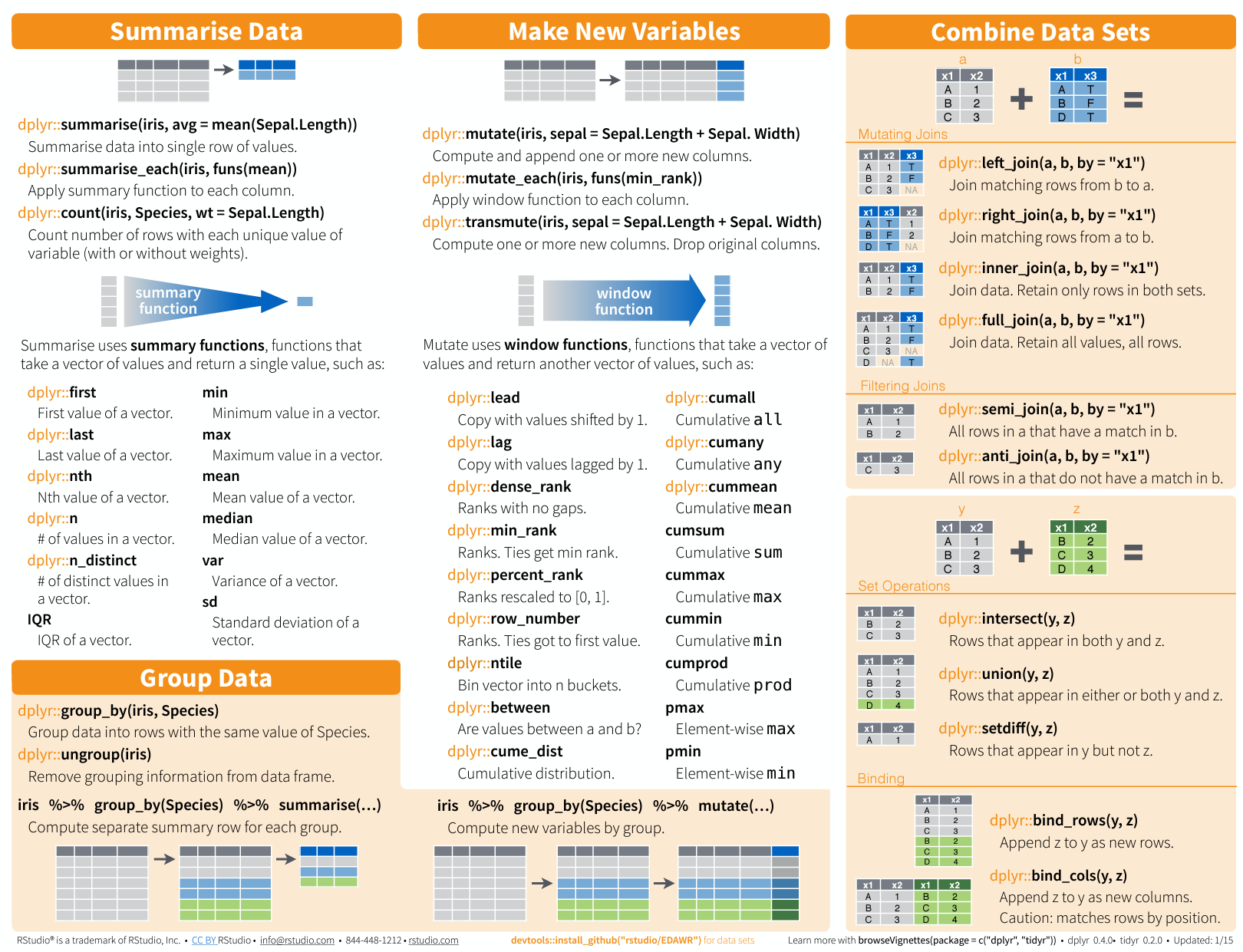
Please see this cheat sheet:
https://www.rstudio.com/wp-content/uploads/2015/02/data-wrangling-cheatsheet.pdf
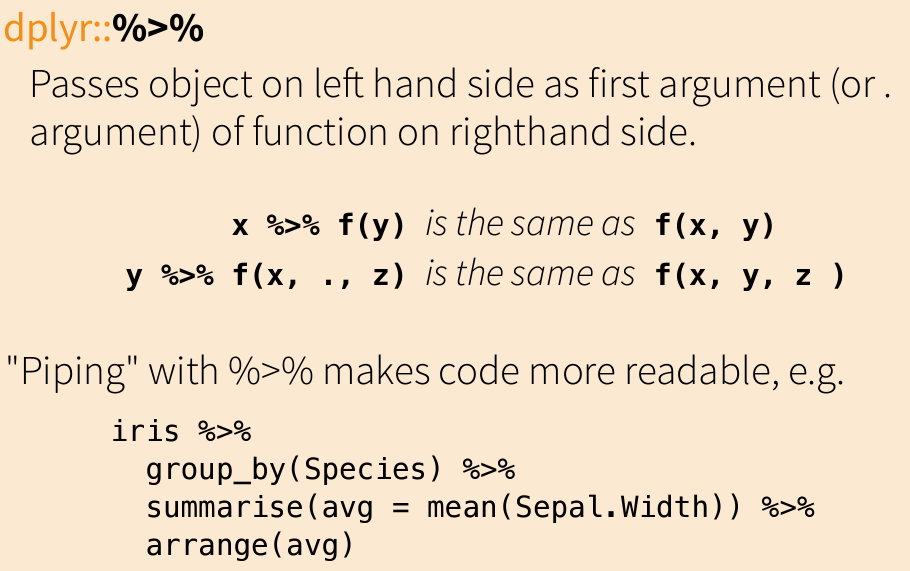
Pipe
Subset Observations
# filter %>% filter (Sepal.Length > 7 )
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 7.1 3.0 5.9 2.1 virginica
2 7.6 3.0 6.6 2.1 virginica
3 7.3 2.9 6.3 1.8 virginica
4 7.2 3.6 6.1 2.5 virginica
5 7.7 3.8 6.7 2.2 virginica
6 7.7 2.6 6.9 2.3 virginica
7 7.7 2.8 6.7 2.0 virginica
8 7.2 3.2 6.0 1.8 virginica
9 7.2 3.0 5.8 1.6 virginica
10 7.4 2.8 6.1 1.9 virginica
11 7.9 3.8 6.4 2.0 virginica
12 7.7 3.0 6.1 2.3 virginica
[1] "setosa" "versicolor" "virginica"
%>% filter (Species == "setosa" )
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5.0 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
11 5.4 3.7 1.5 0.2 setosa
12 4.8 3.4 1.6 0.2 setosa
13 4.8 3.0 1.4 0.1 setosa
14 4.3 3.0 1.1 0.1 setosa
15 5.8 4.0 1.2 0.2 setosa
16 5.7 4.4 1.5 0.4 setosa
17 5.4 3.9 1.3 0.4 setosa
18 5.1 3.5 1.4 0.3 setosa
19 5.7 3.8 1.7 0.3 setosa
20 5.1 3.8 1.5 0.3 setosa
21 5.4 3.4 1.7 0.2 setosa
22 5.1 3.7 1.5 0.4 setosa
23 4.6 3.6 1.0 0.2 setosa
24 5.1 3.3 1.7 0.5 setosa
25 4.8 3.4 1.9 0.2 setosa
26 5.0 3.0 1.6 0.2 setosa
27 5.0 3.4 1.6 0.4 setosa
28 5.2 3.5 1.5 0.2 setosa
29 5.2 3.4 1.4 0.2 setosa
30 4.7 3.2 1.6 0.2 setosa
31 4.8 3.1 1.6 0.2 setosa
32 5.4 3.4 1.5 0.4 setosa
33 5.2 4.1 1.5 0.1 setosa
34 5.5 4.2 1.4 0.2 setosa
35 4.9 3.1 1.5 0.2 setosa
36 5.0 3.2 1.2 0.2 setosa
37 5.5 3.5 1.3 0.2 setosa
38 4.9 3.6 1.4 0.1 setosa
39 4.4 3.0 1.3 0.2 setosa
40 5.1 3.4 1.5 0.2 setosa
41 5.0 3.5 1.3 0.3 setosa
42 4.5 2.3 1.3 0.3 setosa
43 4.4 3.2 1.3 0.2 setosa
44 5.0 3.5 1.6 0.6 setosa
45 5.1 3.8 1.9 0.4 setosa
46 4.8 3.0 1.4 0.3 setosa
47 5.1 3.8 1.6 0.2 setosa
48 4.6 3.2 1.4 0.2 setosa
49 5.3 3.7 1.5 0.2 setosa
50 5.0 3.3 1.4 0.2 setosa
%>% filter (Species == "setosa" ) %>% plot%>% filter (Species == "setosa" ) %>% summary
Sepal.Length Sepal.Width Petal.Length Petal.Width
Min. :4.300 Min. :2.300 Min. :1.000 Min. :0.100
1st Qu.:4.800 1st Qu.:3.200 1st Qu.:1.400 1st Qu.:0.200
Median :5.000 Median :3.400 Median :1.500 Median :0.200
Mean :5.006 Mean :3.428 Mean :1.462 Mean :0.246
3rd Qu.:5.200 3rd Qu.:3.675 3rd Qu.:1.575 3rd Qu.:0.300
Max. :5.800 Max. :4.400 Max. :1.900 Max. :0.600
Species
setosa :50
versicolor: 0
virginica : 0
# distinct: remove duplication (take only unique values) %>% distinct (Species)
Species
1 setosa
2 versicolor
3 virginica
# random sampling %>% nrow%>% sample_frac (0.5 , replace= T)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.2 4.1 1.5 0.1 setosa
2 6.5 3.2 5.1 2.0 virginica
3 6.5 2.8 4.6 1.5 versicolor
4 6.1 3.0 4.6 1.4 versicolor
5 5.7 2.6 3.5 1.0 versicolor
6 6.5 3.0 5.5 1.8 virginica
7 7.2 3.6 6.1 2.5 virginica
8 6.3 2.5 5.0 1.9 virginica
9 4.3 3.0 1.1 0.1 setosa
10 4.6 3.6 1.0 0.2 setosa
11 5.6 2.9 3.6 1.3 versicolor
12 7.6 3.0 6.6 2.1 virginica
13 6.3 2.5 4.9 1.5 versicolor
14 4.6 3.6 1.0 0.2 setosa
15 7.4 2.8 6.1 1.9 virginica
16 6.4 2.8 5.6 2.1 virginica
17 5.5 2.6 4.4 1.2 versicolor
18 7.7 3.0 6.1 2.3 virginica
19 6.4 3.1 5.5 1.8 virginica
20 6.3 2.5 4.9 1.5 versicolor
21 6.2 2.2 4.5 1.5 versicolor
22 5.8 2.7 4.1 1.0 versicolor
23 6.9 3.2 5.7 2.3 virginica
24 6.7 2.5 5.8 1.8 virginica
25 4.8 3.4 1.6 0.2 setosa
26 5.7 4.4 1.5 0.4 setosa
27 5.8 4.0 1.2 0.2 setosa
28 7.7 2.6 6.9 2.3 virginica
29 6.9 3.1 5.1 2.3 virginica
30 7.7 3.0 6.1 2.3 virginica
31 4.8 3.4 1.9 0.2 setosa
32 6.8 3.2 5.9 2.3 virginica
33 5.1 2.5 3.0 1.1 versicolor
34 7.9 3.8 6.4 2.0 virginica
35 6.3 3.4 5.6 2.4 virginica
36 4.9 3.1 1.5 0.2 setosa
37 5.9 3.0 4.2 1.5 versicolor
38 6.1 2.8 4.7 1.2 versicolor
39 5.1 3.8 1.9 0.4 setosa
40 6.9 3.1 4.9 1.5 versicolor
41 5.4 3.4 1.5 0.4 setosa
42 6.3 2.7 4.9 1.8 virginica
43 5.2 2.7 3.9 1.4 versicolor
44 5.7 3.0 4.2 1.2 versicolor
45 5.5 2.5 4.0 1.3 versicolor
46 6.7 3.3 5.7 2.1 virginica
47 5.6 2.5 3.9 1.1 versicolor
48 4.9 3.1 1.5 0.2 setosa
49 6.1 2.9 4.7 1.4 versicolor
50 6.5 2.8 4.6 1.5 versicolor
51 5.4 3.0 4.5 1.5 versicolor
52 5.0 3.3 1.4 0.2 setosa
53 4.4 3.2 1.3 0.2 setosa
54 6.4 2.8 5.6 2.1 virginica
55 5.0 2.0 3.5 1.0 versicolor
56 5.5 2.4 3.8 1.1 versicolor
57 6.3 3.3 4.7 1.6 versicolor
58 4.9 3.0 1.4 0.2 setosa
59 5.0 3.5 1.3 0.3 setosa
60 6.2 2.8 4.8 1.8 virginica
61 6.8 3.2 5.9 2.3 virginica
62 5.1 3.8 1.6 0.2 setosa
63 4.8 3.0 1.4 0.1 setosa
64 6.3 2.8 5.1 1.5 virginica
65 5.7 2.8 4.1 1.3 versicolor
66 6.6 3.0 4.4 1.4 versicolor
67 6.7 3.1 4.7 1.5 versicolor
68 7.2 3.2 6.0 1.8 virginica
69 6.6 3.0 4.4 1.4 versicolor
70 7.7 2.6 6.9 2.3 virginica
71 6.7 3.1 4.7 1.5 versicolor
72 7.2 3.0 5.8 1.6 virginica
73 6.9 3.1 5.1 2.3 virginica
74 6.7 3.1 5.6 2.4 virginica
75 7.6 3.0 6.6 2.1 virginica
%>% sample_frac (0.5 , replace= T) %>% nrow%>% sample_n (10 , replace= T)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.6 3.0 4.5 1.5 versicolor
2 6.8 3.2 5.9 2.3 virginica
3 5.6 2.5 3.9 1.1 versicolor
4 6.3 2.9 5.6 1.8 virginica
5 6.3 2.3 4.4 1.3 versicolor
6 6.0 3.0 4.8 1.8 virginica
7 5.1 3.8 1.6 0.2 setosa
8 5.2 3.5 1.5 0.2 setosa
9 7.6 3.0 6.6 2.1 virginica
10 5.1 3.5 1.4 0.2 setosa
%>% sample_n (10 , replace= T) %>% nrow # slice %>% slice (10 : 15 )
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 4.9 3.1 1.5 0.1 setosa
2 5.4 3.7 1.5 0.2 setosa
3 4.8 3.4 1.6 0.2 setosa
4 4.8 3.0 1.4 0.1 setosa
5 4.3 3.0 1.1 0.1 setosa
6 5.8 4.0 1.2 0.2 setosa
# Top n in X %>% top_n (5 , Sepal.Length)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 7.7 3.8 6.7 2.2 virginica
2 7.7 2.6 6.9 2.3 virginica
3 7.7 2.8 6.7 2.0 virginica
4 7.9 3.8 6.4 2.0 virginica
5 7.7 3.0 6.1 2.3 virginica
%>% top_n (5 , Sepal.Width)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.4 3.9 1.7 0.4 setosa
2 5.8 4.0 1.2 0.2 setosa
3 5.7 4.4 1.5 0.4 setosa
4 5.4 3.9 1.3 0.4 setosa
5 5.2 4.1 1.5 0.1 setosa
6 5.5 4.2 1.4 0.2 setosa
Subset Variables
# pull & select %>% pull (Petal.Width)
[1] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 0.2 0.2 0.1 0.1 0.2 0.4 0.4 0.3
[19] 0.3 0.3 0.2 0.4 0.2 0.5 0.2 0.2 0.4 0.2 0.2 0.2 0.2 0.4 0.1 0.2 0.2 0.2
[37] 0.2 0.1 0.2 0.2 0.3 0.3 0.2 0.6 0.4 0.3 0.2 0.2 0.2 0.2 1.4 1.5 1.5 1.3
[55] 1.5 1.3 1.6 1.0 1.3 1.4 1.0 1.5 1.0 1.4 1.3 1.4 1.5 1.0 1.5 1.1 1.8 1.3
[73] 1.5 1.2 1.3 1.4 1.4 1.7 1.5 1.0 1.1 1.0 1.2 1.6 1.5 1.6 1.5 1.3 1.3 1.3
[91] 1.2 1.4 1.2 1.0 1.3 1.2 1.3 1.3 1.1 1.3 2.5 1.9 2.1 1.8 2.2 2.1 1.7 1.8
[109] 1.8 2.5 2.0 1.9 2.1 2.0 2.4 2.3 1.8 2.2 2.3 1.5 2.3 2.0 2.0 1.8 2.1 1.8
[127] 1.8 1.8 2.1 1.6 1.9 2.0 2.2 1.5 1.4 2.3 2.4 1.8 1.8 2.1 2.4 2.3 1.9 2.3
[145] 2.5 2.3 1.9 2.0 2.3 1.8
%>% select (Petal.Width)
Petal.Width
1 0.2
2 0.2
3 0.2
4 0.2
5 0.2
6 0.4
7 0.3
8 0.2
9 0.2
10 0.1
11 0.2
12 0.2
13 0.1
14 0.1
15 0.2
16 0.4
17 0.4
18 0.3
19 0.3
20 0.3
21 0.2
22 0.4
23 0.2
24 0.5
25 0.2
26 0.2
27 0.4
28 0.2
29 0.2
30 0.2
31 0.2
32 0.4
33 0.1
34 0.2
35 0.2
36 0.2
37 0.2
38 0.1
39 0.2
40 0.2
41 0.3
42 0.3
43 0.2
44 0.6
45 0.4
46 0.3
47 0.2
48 0.2
49 0.2
50 0.2
51 1.4
52 1.5
53 1.5
54 1.3
55 1.5
56 1.3
57 1.6
58 1.0
59 1.3
60 1.4
61 1.0
62 1.5
63 1.0
64 1.4
65 1.3
66 1.4
67 1.5
68 1.0
69 1.5
70 1.1
71 1.8
72 1.3
73 1.5
74 1.2
75 1.3
76 1.4
77 1.4
78 1.7
79 1.5
80 1.0
81 1.1
82 1.0
83 1.2
84 1.6
85 1.5
86 1.6
87 1.5
88 1.3
89 1.3
90 1.3
91 1.2
92 1.4
93 1.2
94 1.0
95 1.3
96 1.2
97 1.3
98 1.3
99 1.1
100 1.3
101 2.5
102 1.9
103 2.1
104 1.8
105 2.2
106 2.1
107 1.7
108 1.8
109 1.8
110 2.5
111 2.0
112 1.9
113 2.1
114 2.0
115 2.4
116 2.3
117 1.8
118 2.2
119 2.3
120 1.5
121 2.3
122 2.0
123 2.0
124 1.8
125 2.1
126 1.8
127 1.8
128 1.8
129 2.1
130 1.6
131 1.9
132 2.0
133 2.2
134 1.5
135 1.4
136 2.3
137 2.4
138 1.8
139 1.8
140 2.1
141 2.4
142 2.3
143 1.9
144 2.3
145 2.5
146 2.3
147 1.9
148 2.0
149 2.3
150 1.8
%>% pull (Petal.Width) %>% str
num [1:150] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
%>% select (Petal.Width) %>% str
'data.frame': 150 obs. of 1 variable:
$ Petal.Width: num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
%>% select (Petal.Length, Petal.Width) %>% head
Petal.Length Petal.Width
1 1.4 0.2
2 1.4 0.2
3 1.3 0.2
4 1.5 0.2
5 1.4 0.2
6 1.7 0.4
# useful helpers: starts_with(), contains() %>% select (starts_with ("Peta" )) %>% head
Petal.Length Petal.Width
1 1.4 0.2
2 1.4 0.2
3 1.3 0.2
4 1.5 0.2
5 1.4 0.2
6 1.7 0.4
%>% select (contains ("tal" )) %>% head
Petal.Length Petal.Width
1 1.4 0.2
2 1.4 0.2
3 1.3 0.2
4 1.5 0.2
5 1.4 0.2
6 1.7 0.4
Reshaping & Arrange data
# let's use mtcars dataset
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
# About mtcars dataset # help(mtcars) # arrange %>% arrange (mpg) %>% head
mpg cyl disp hp drat wt qsec vs am gear carb
Cadillac Fleetwood 10.4 8 472 205 2.93 5.250 17.98 0 0 3 4
Lincoln Continental 10.4 8 460 215 3.00 5.424 17.82 0 0 3 4
Camaro Z28 13.3 8 350 245 3.73 3.840 15.41 0 0 3 4
Duster 360 14.3 8 360 245 3.21 3.570 15.84 0 0 3 4
Chrysler Imperial 14.7 8 440 230 3.23 5.345 17.42 0 0 3 4
Maserati Bora 15.0 8 301 335 3.54 3.570 14.60 0 1 5 8
%>% add_rownames %>% head
Warning: `add_rownames()` was deprecated in dplyr 1.0.0.
ℹ Please use `tibble::rownames_to_column()` instead.
# A tibble: 6 × 12
rowname mpg cyl disp hp drat wt qsec vs am gear carb
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Mazda RX4 21 6 160 110 3.9 2.62 16.5 0 1 4 4
2 Mazda RX4 W… 21 6 160 110 3.9 2.88 17.0 0 1 4 4
3 Datsun 710 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
4 Hornet 4 Dr… 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
5 Hornet Spor… 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
6 Valiant 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
%>% add_rownames %>% arrange (mpg) %>%
Warning: `add_rownames()` was deprecated in dplyr 1.0.0.
ℹ Please use `tibble::rownames_to_column()` instead.
# A tibble: 6 × 12
rowname mpg cyl disp hp drat wt qsec vs am gear carb
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Cadillac Fl… 10.4 8 472 205 2.93 5.25 18.0 0 0 3 4
2 Lincoln Con… 10.4 8 460 215 3 5.42 17.8 0 0 3 4
3 Camaro Z28 13.3 8 350 245 3.73 3.84 15.4 0 0 3 4
4 Duster 360 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4
5 Chrysler Im… 14.7 8 440 230 3.23 5.34 17.4 0 0 3 4
6 Maserati Bo… 15 8 301 335 3.54 3.57 14.6 0 1 5 8
%>% add_rownames %>% arrange (desc (mpg)) %>% select (rowname)
Warning: `add_rownames()` was deprecated in dplyr 1.0.0.
ℹ Please use `tibble::rownames_to_column()` instead.
# A tibble: 32 × 1
rowname
<chr>
1 Toyota Corolla
2 Fiat 128
3 Honda Civic
4 Lotus Europa
5 Fiat X1-9
6 Porsche 914-2
7 Merc 240D
8 Datsun 710
9 Merc 230
10 Toyota Corona
# ℹ 22 more rows
Summarise data
# Summarise %>% summarise (avg.PL= mean (Petal.Length))%>% summarise (sd.PL= sd (Petal.Length))%>% summarise (avg.PL= mean (Petal.Length),sd.PL= sd (Petal.Length),min.PL= min (Petal.Length))
avg.PL sd.PL min.PL
1 3.758 1.765298 1
Species n
1 setosa 50
2 versicolor 50
3 virginica 50
%>% sample_frac (0.3 ) %>% count (Species)
Species n
1 setosa 22
2 versicolor 12
3 virginica 11
%>% summarise_all (mean)
Warning: There was 1 warning in `summarise()`.
ℹ In argument: `Species = (function (x, ...) ...`.
Caused by warning in `mean.default()`:
! argument is not numeric or logical: returning NA
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.843333 3.057333 3.758 1.199333 NA
%>% summarise_at ("Petal.Length" , sum)%>% summarise_at (c ("Petal.Length" , "Petal.Width" ), mean)
Petal.Length Petal.Width
1 3.758 1.199333
Group and Summarise data
# Group %>% group_by (Species)
# A tibble: 150 × 5
# Groups: Species [3]
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rows
%>% group_by (Species) %>% summarise_all (mean)
# A tibble: 3 × 5
Species Sepal.Length Sepal.Width Petal.Length Petal.Width
<fct> <dbl> <dbl> <dbl> <dbl>
1 setosa 5.01 3.43 1.46 0.246
2 versicolor 5.94 2.77 4.26 1.33
3 virginica 6.59 2.97 5.55 2.03
%>% group_by (Species) %>% select (starts_with ("Sep" )) %>% summarise_all (mean)
Adding missing grouping variables: `Species`
# A tibble: 3 × 3
Species Sepal.Length Sepal.Width
<fct> <dbl> <dbl>
1 setosa 5.01 3.43
2 versicolor 5.94 2.77
3 virginica 6.59 2.97
%>% group_by (am) %>% summarise (hp.avg= mean (hp),hp.sd= sd (hp))
# A tibble: 2 × 3
am hp.avg hp.sd
<dbl> <dbl> <dbl>
1 0 160. 53.9
2 1 127. 84.1
Make New Variables
# mutate # mpg to kml %>% mutate (kml= 0.425144 * mpg)
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
kml
Mazda RX4 8.928024
Mazda RX4 Wag 8.928024
Datsun 710 9.693283
Hornet 4 Drive 9.098082
Hornet Sportabout 7.950193
Valiant 7.695106
Duster 360 6.079559
Merc 240D 10.373514
Merc 230 9.693283
Merc 280 8.162765
Merc 280C 7.567563
Merc 450SE 6.972362
Merc 450SL 7.354991
Merc 450SLC 6.462189
Cadillac Fleetwood 4.421498
Lincoln Continental 4.421498
Chrysler Imperial 6.249617
Fiat 128 13.774666
Honda Civic 12.924378
Toyota Corolla 14.412382
Toyota Corona 9.140596
Dodge Challenger 6.589732
AMC Javelin 6.462189
Camaro Z28 5.654415
Pontiac Firebird 8.162765
Fiat X1-9 11.606431
Porsche 914-2 11.053744
Lotus Europa 12.924378
Ford Pantera L 6.717275
Ferrari Dino 8.375337
Maserati Bora 6.377160
Volvo 142E 9.098082
%>% mutate (kml= round (0.425144 * mpg, 1 ))
mpg cyl disp hp drat wt qsec vs am gear carb kml
Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 8.9
Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 8.9
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 9.7
Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 9.1
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 8.0
Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1 7.7
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 6.1
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 10.4
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2 9.7
Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4 8.2
Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4 7.6
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3 7.0
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3 7.4
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3 6.5
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 4.4
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4 4.4
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4 6.2
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 13.8
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2 12.9
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1 14.4
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1 9.1
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2 6.6
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2 6.5
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4 5.7
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2 8.2
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1 11.6
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2 11.1
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2 12.9
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4 6.7
Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6 8.4
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8 6.4
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2 9.1
%>% mutate (kml= round (0.425144 * mpg, 1 )) %>% select (mpg, kml) %>% top_n (5 , mpg)
mpg kml
Fiat 128 32.4 13.8
Honda Civic 30.4 12.9
Toyota Corolla 33.9 14.4
Fiat X1-9 27.3 11.6
Lotus Europa 30.4 12.9
# transmute %>% transmute (hp/ wt)
hp/wt
Mazda RX4 41.98473
Mazda RX4 Wag 38.26087
Datsun 710 40.08621
Hornet 4 Drive 34.21462
Hornet Sportabout 50.87209
Valiant 30.34682
Duster 360 68.62745
Merc 240D 19.43574
Merc 230 30.15873
Merc 280 35.75581
Merc 280C 35.75581
Merc 450SE 44.22604
Merc 450SL 48.25737
Merc 450SLC 47.61905
Cadillac Fleetwood 39.04762
Lincoln Continental 39.63864
Chrysler Imperial 43.03087
Fiat 128 30.00000
Honda Civic 32.19814
Toyota Corolla 35.42234
Toyota Corona 39.35091
Dodge Challenger 42.61364
AMC Javelin 43.66812
Camaro Z28 63.80208
Pontiac Firebird 45.51365
Fiat X1-9 34.10853
Porsche 914-2 42.52336
Lotus Europa 74.68605
Ford Pantera L 83.28076
Ferrari Dino 63.17690
Maserati Bora 93.83754
Volvo 142E 39.20863
# rename %>% names
[1] "mpg" "cyl" "disp" "hp" "drat" "wt" "qsec" "vs" "am" "gear"
[11] "carb"
%>% mutate (new= 1 ) %>% head
mpg cyl disp hp drat wt qsec vs am gear carb new
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 1
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 1
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 1
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 1
%>% mutate (new= 1 ) %>% rename (change.name= new) %>% head
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
change.name
Mazda RX4 1
Mazda RX4 Wag 1
Datsun 710 1
Hornet 4 Drive 1
Hornet Sportabout 1
Valiant 1
Combine Data Sets
Joins!
Thanks to Garrick Aden-Buie (garrickadenbuie.com ), we can teach joins very easily with his fantastic diagrams.
Let’s use some example datasets that come pre-loaded in R to demonstrate how joins work using dplyrmtcars
First, let’s set up our example data frames from the mtcars
# Create two data frames from mtcars <- mtcars[1 : 10 , c ("mpg" , "cyl" , "disp" )] # first 10 rows, select specific columns <- mtcars[5 : 15 , c ("disp" , "hp" , "drat" )] # rows 5 to 15, select specific columns # Make sure there's a common key for joining; here, we'll use 'disp' # Print out the data frames to see what they contain print (df1)
mpg cyl disp
Mazda RX4 21.0 6 160.0
Mazda RX4 Wag 21.0 6 160.0
Datsun 710 22.8 4 108.0
Hornet 4 Drive 21.4 6 258.0
Hornet Sportabout 18.7 8 360.0
Valiant 18.1 6 225.0
Duster 360 14.3 8 360.0
Merc 240D 24.4 4 146.7
Merc 230 22.8 4 140.8
Merc 280 19.2 6 167.6
disp hp drat
Hornet Sportabout 360.0 175 3.15
Valiant 225.0 105 2.76
Duster 360 360.0 245 3.21
Merc 240D 146.7 62 3.69
Merc 230 140.8 95 3.92
Merc 280 167.6 123 3.92
Merc 280C 167.6 123 3.92
Merc 450SE 275.8 180 3.07
Merc 450SL 275.8 180 3.07
Merc 450SLC 275.8 180 3.07
Cadillac Fleetwood 472.0 205 2.93
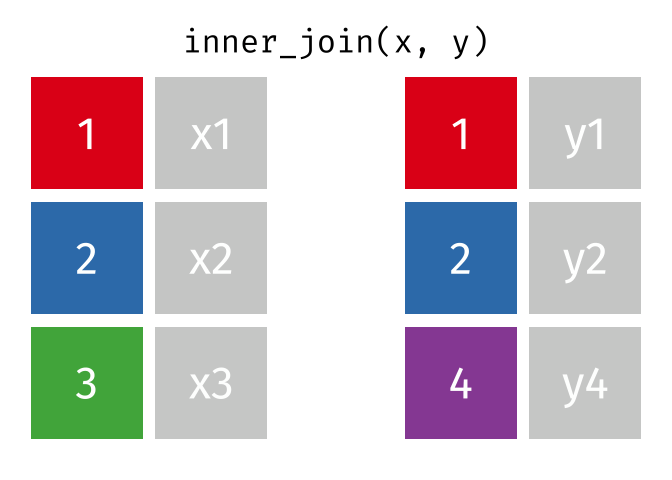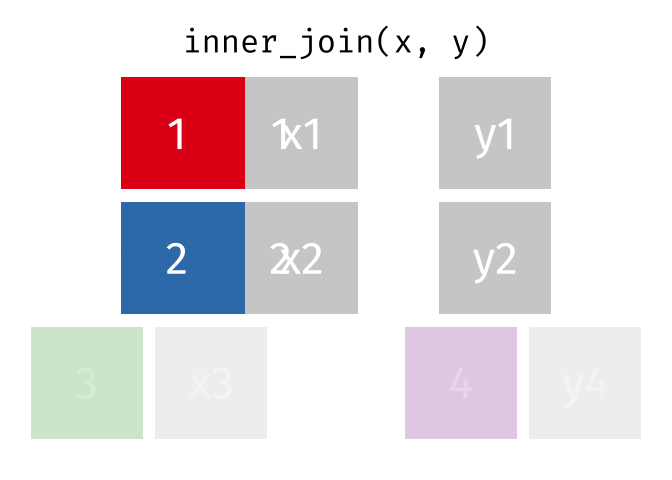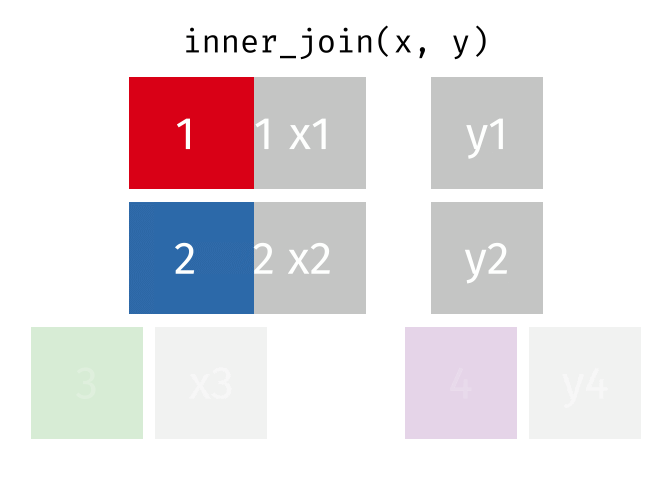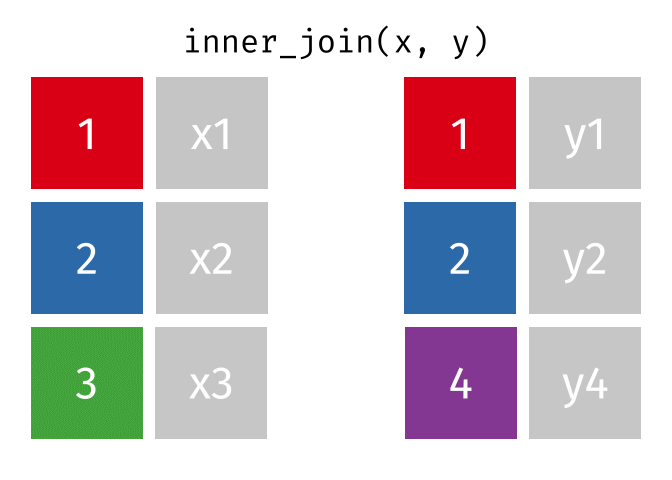
Inner Join
All rows from x where there are matching values in y, and all columns from x and y.
<- inner_join (df1, df2, by = "disp" )
Warning in inner_join(df1, df2, by = "disp"): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 5 of `x` matches multiple rows in `y`.
ℹ Row 1 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.
mpg cyl disp hp drat
1 18.7 8 360.0 175 3.15
2 18.7 8 360.0 245 3.21
3 18.1 6 225.0 105 2.76
4 14.3 8 360.0 175 3.15
5 14.3 8 360.0 245 3.21
6 24.4 4 146.7 62 3.69
7 22.8 4 140.8 95 3.92
8 19.2 6 167.6 123 3.92
9 19.2 6 167.6 123 3.92
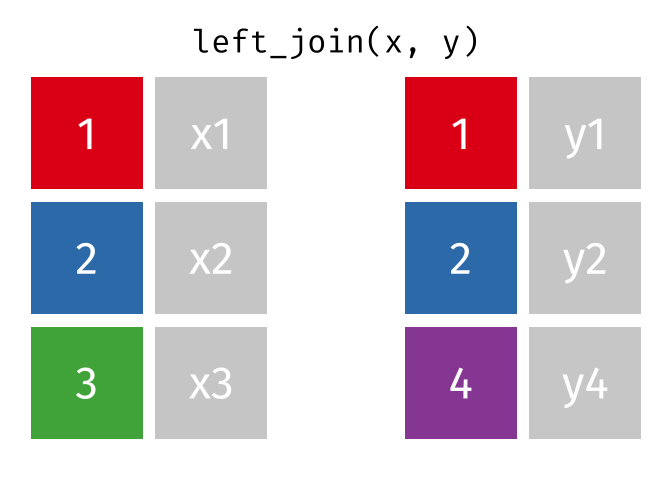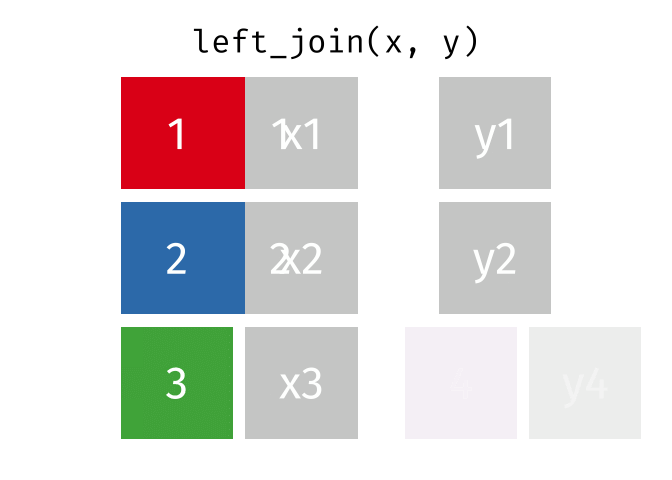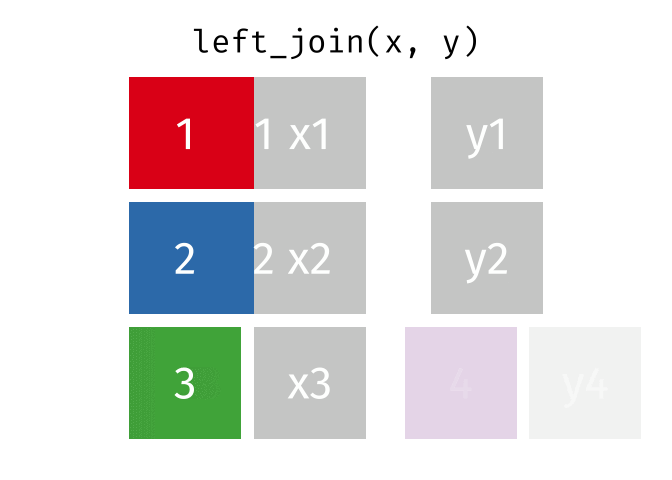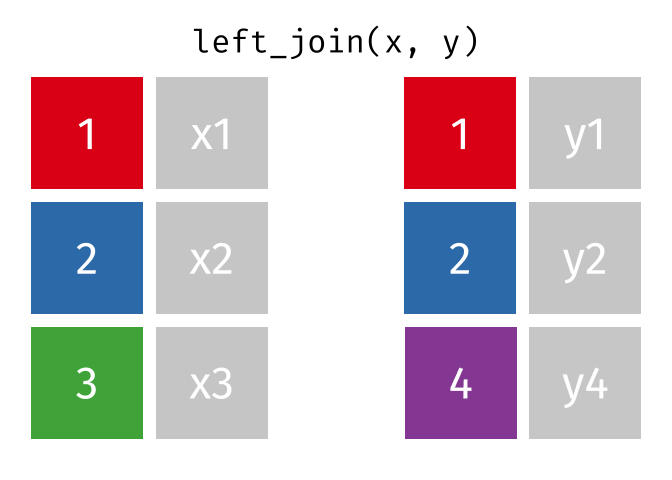
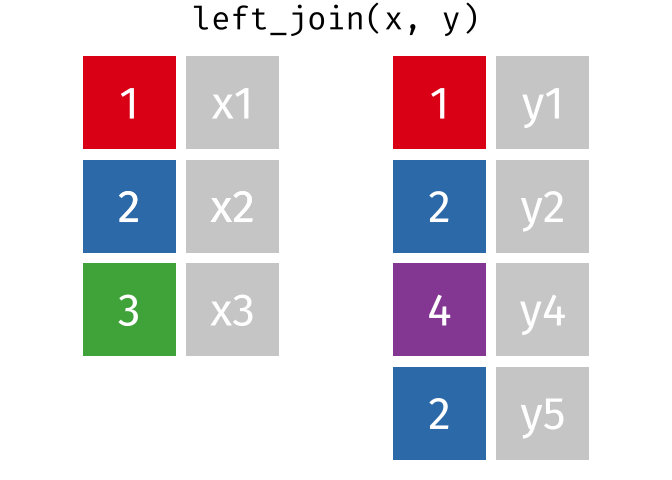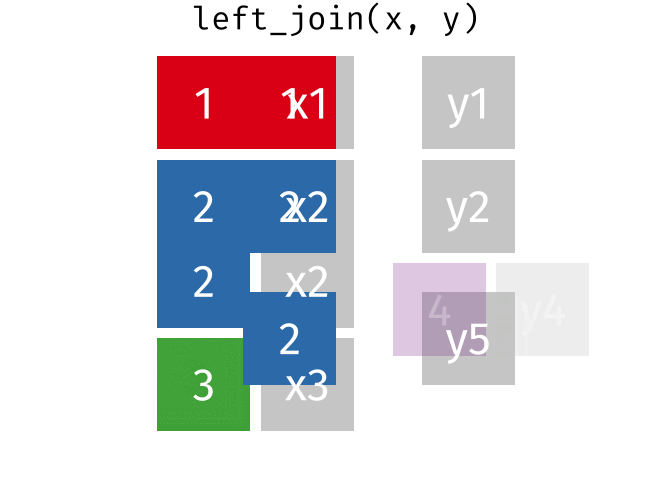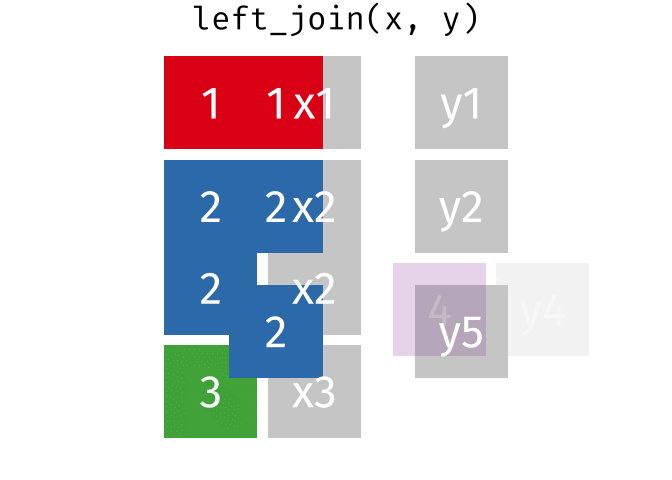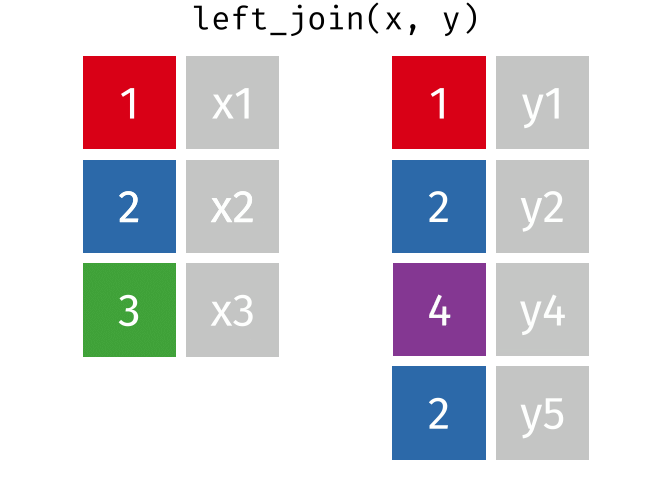
Left Join
All rows from x, and all columns from x and y. Rows in x with no match in y will have NA values in the new columns.
<- left_join (df1, df2, by = "disp" )
Warning in left_join(df1, df2, by = "disp"): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 5 of `x` matches multiple rows in `y`.
ℹ Row 1 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.
mpg cyl disp hp drat
1 21.0 6 160.0 NA NA
2 21.0 6 160.0 NA NA
3 22.8 4 108.0 NA NA
4 21.4 6 258.0 NA NA
5 18.7 8 360.0 175 3.15
6 18.7 8 360.0 245 3.21
7 18.1 6 225.0 105 2.76
8 14.3 8 360.0 175 3.15
9 14.3 8 360.0 245 3.21
10 24.4 4 146.7 62 3.69
11 22.8 4 140.8 95 3.92
12 19.2 6 167.6 123 3.92
13 19.2 6 167.6 123 3.92
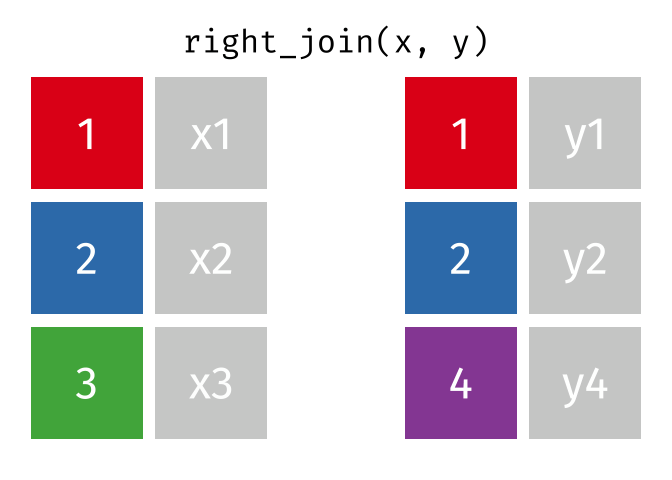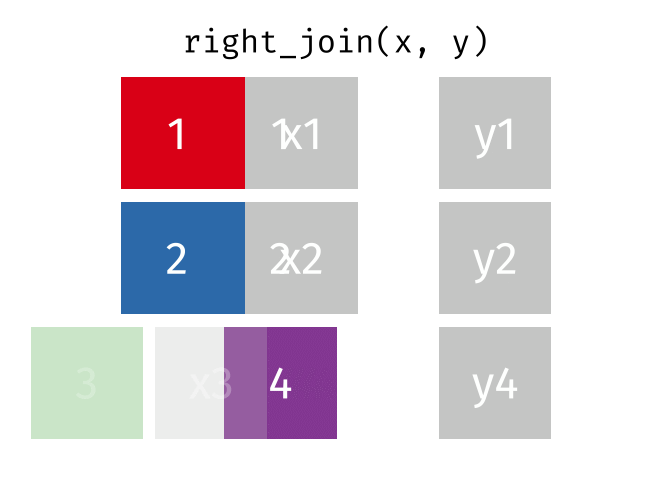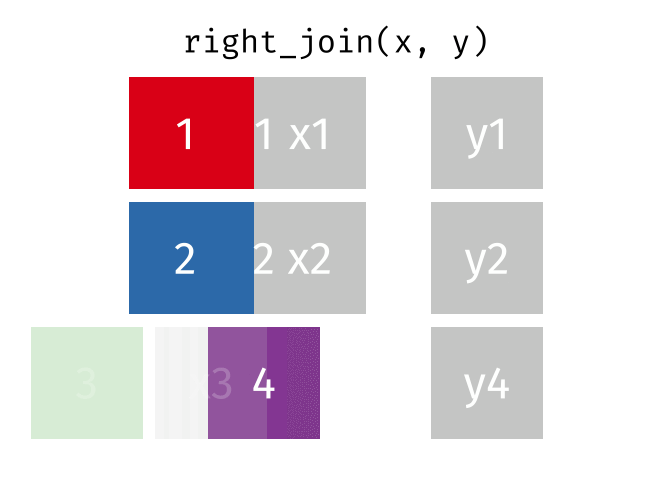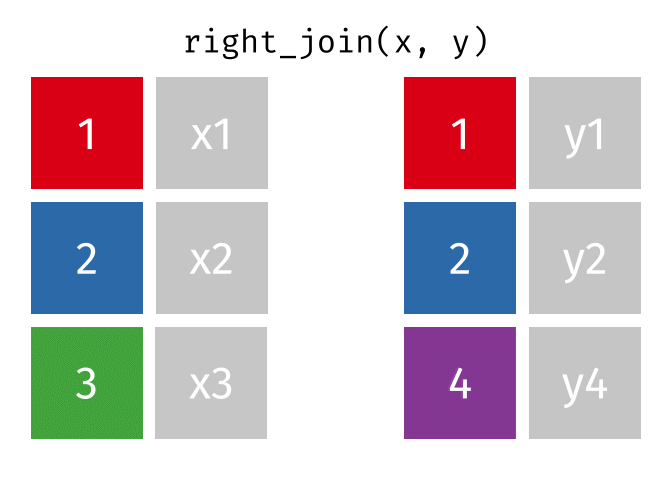
Right Join
All rows from y, and all columns from x and y. Rows in y with no match in x will have NA values in the new columns.
<- right_join (df1, df2, by = "disp" )
Warning in right_join(df1, df2, by = "disp"): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 5 of `x` matches multiple rows in `y`.
ℹ Row 1 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.
mpg cyl disp hp drat
1 18.7 8 360.0 175 3.15
2 18.7 8 360.0 245 3.21
3 18.1 6 225.0 105 2.76
4 14.3 8 360.0 175 3.15
5 14.3 8 360.0 245 3.21
6 24.4 4 146.7 62 3.69
7 22.8 4 140.8 95 3.92
8 19.2 6 167.6 123 3.92
9 19.2 6 167.6 123 3.92
10 NA NA 275.8 180 3.07
11 NA NA 275.8 180 3.07
12 NA NA 275.8 180 3.07
13 NA NA 472.0 205 2.93
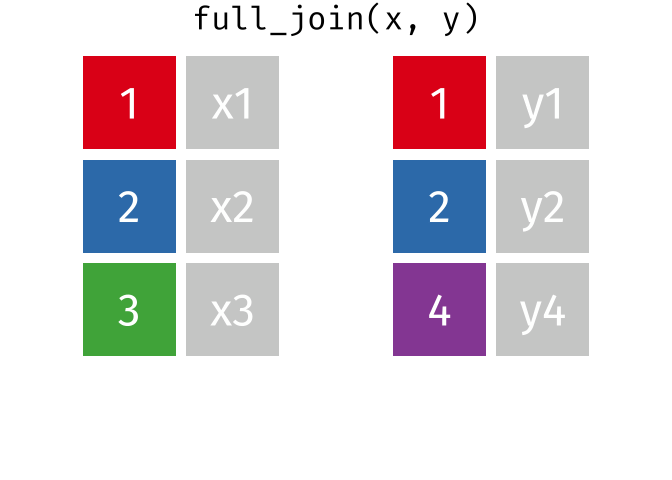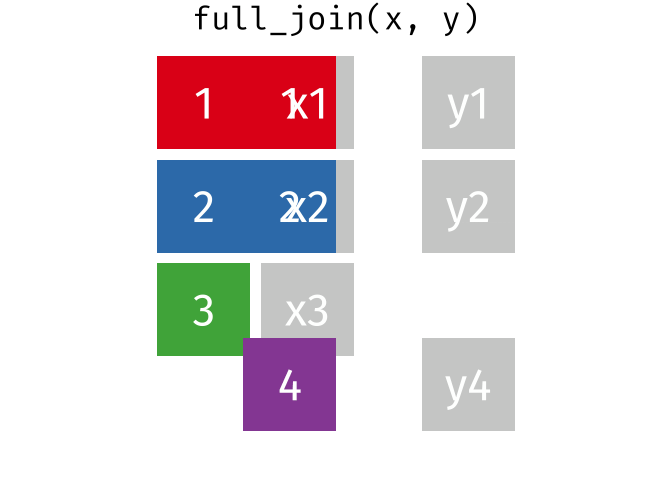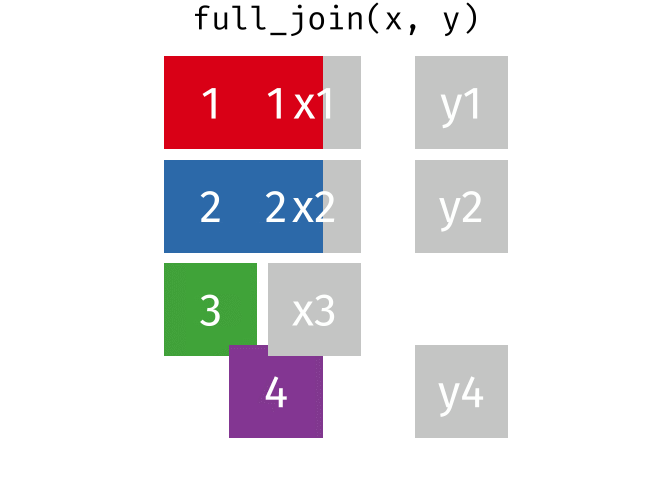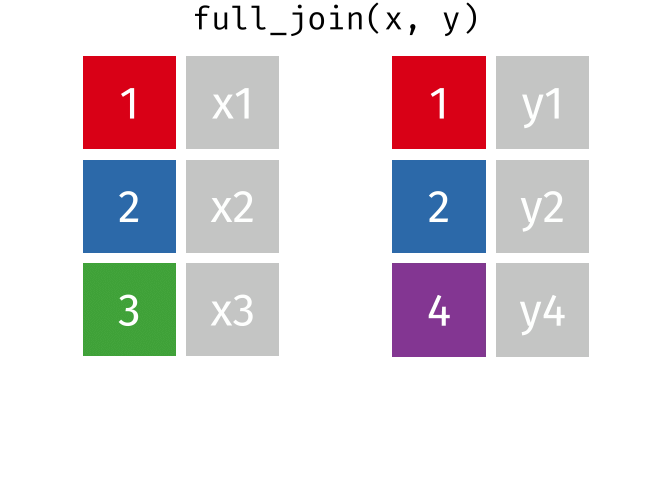
Full Join
All rows and all columns from both x and y. Where there are not matching values, returns NA for the one missing.
<- full_join (df1, df2, by = "disp" )
Warning in full_join(df1, df2, by = "disp"): Detected an unexpected many-to-many relationship between `x` and `y`.
ℹ Row 5 of `x` matches multiple rows in `y`.
ℹ Row 1 of `y` matches multiple rows in `x`.
ℹ If a many-to-many relationship is expected, set `relationship =
"many-to-many"` to silence this warning.
mpg cyl disp hp drat
1 21.0 6 160.0 NA NA
2 21.0 6 160.0 NA NA
3 22.8 4 108.0 NA NA
4 21.4 6 258.0 NA NA
5 18.7 8 360.0 175 3.15
6 18.7 8 360.0 245 3.21
7 18.1 6 225.0 105 2.76
8 14.3 8 360.0 175 3.15
9 14.3 8 360.0 245 3.21
10 24.4 4 146.7 62 3.69
11 22.8 4 140.8 95 3.92
12 19.2 6 167.6 123 3.92
13 19.2 6 167.6 123 3.92
14 NA NA 275.8 180 3.07
15 NA NA 275.8 180 3.07
16 NA NA 275.8 180 3.07
17 NA NA 472.0 205 2.93
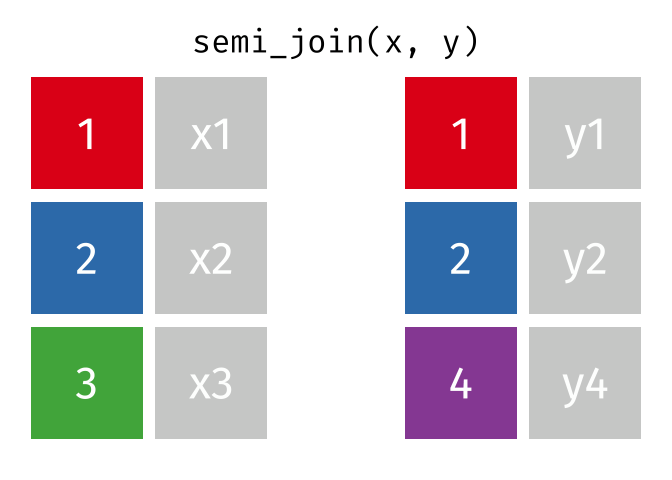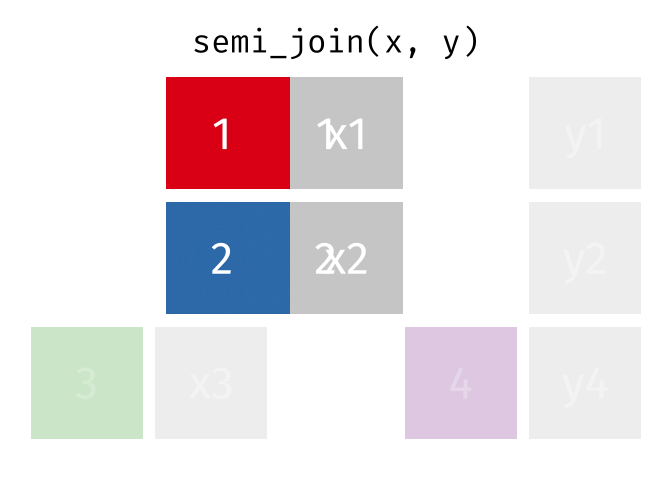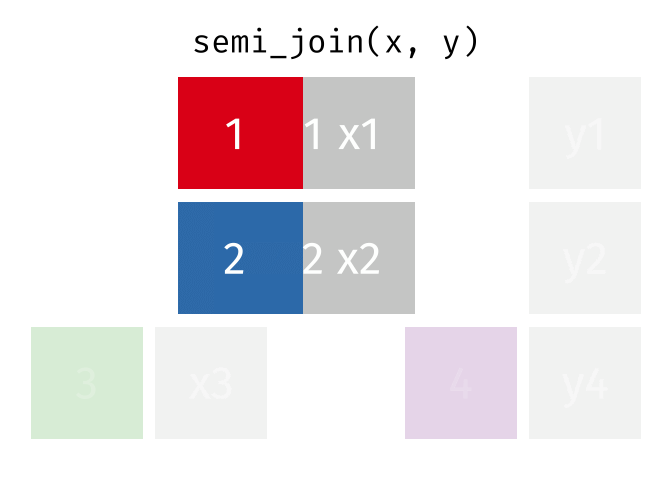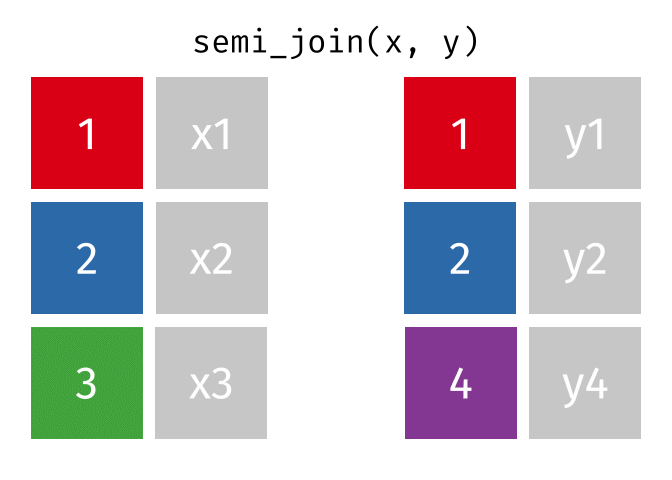
Filtering Joins
Filtering joins match observations in the same way as mutating joins, but affect the observations, not the variables. … Semi-joins are useful for matching filtered summary tables back to the original rows. … Anti-joins are useful for diagnosing join mismatches.R for Data Science: Filtering Joins
Semi Join
All rows from x where there are matching values in y, keeping just columns from x.
<- semi_join (df1, df2, by = "disp" )print (semi_join_result)
mpg cyl disp
Hornet Sportabout 18.7 8 360.0
Valiant 18.1 6 225.0
Duster 360 14.3 8 360.0
Merc 240D 24.4 4 146.7
Merc 230 22.8 4 140.8
Merc 280 19.2 6 167.6
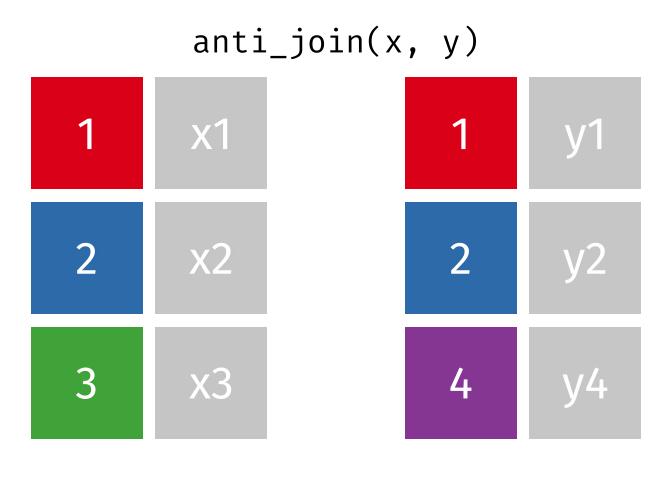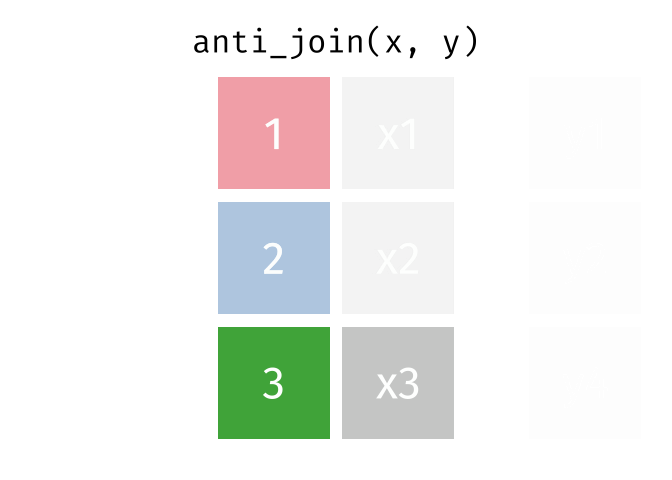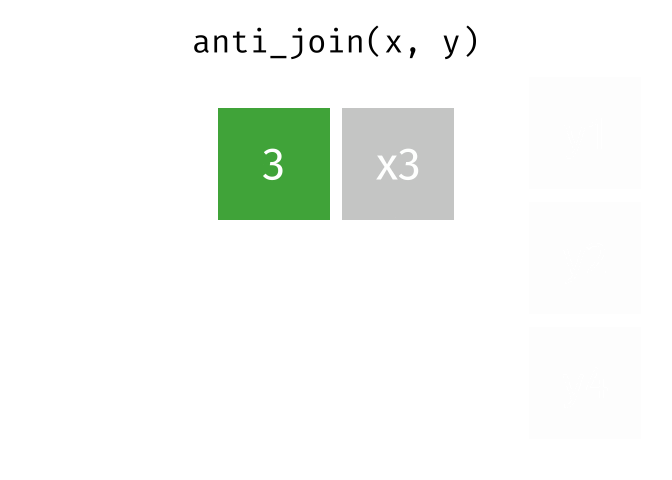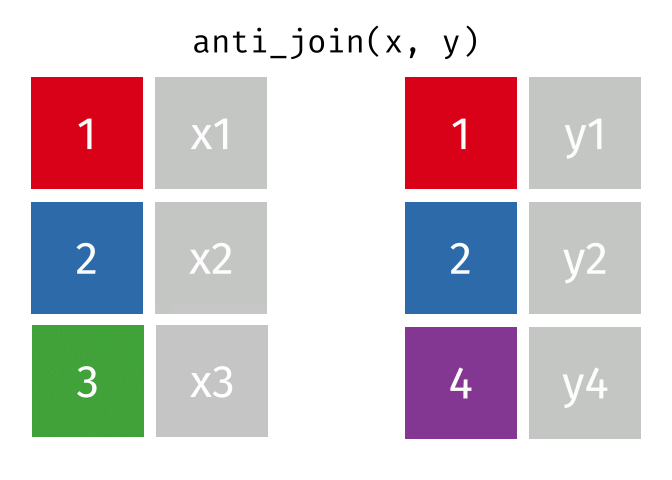
Anti Join
All rows from x where there are not matching values in y, keeping just column
# Assuming df1 and df2 from the previous example <- anti_join (df1, df2, by = "disp" )print (anti_join_result)
mpg cyl disp
Mazda RX4 21.0 6 160
Mazda RX4 Wag 21.0 6 160
Datsun 710 22.8 4 108
Hornet 4 Drive 21.4 6 258